Abstract
Functional regeneration after nervous system injury requires transected axons to reconnect with their original target tissue. Axonal fusion, a spontaneous regenerative mechanism identified in several species, provides an efficient means of achieving target reconnection as a regrowing axon is able to contact and fuse with its own separated axon fragment, thereby re-establishing the original axonal tract1,2,3,4,5,6,7. Here we report a molecular characterization of this process in Caenorhabditis elegans, revealing dynamic changes in the subcellular localization of the EFF-1 fusogen after axotomy, and establishing phosphatidylserine (PS) and the PS receptor (PSR-1) as critical components for axonal fusion. PSR-1 functions cell-autonomously in the regrowing neuron and, instead of acting in its canonical signalling pathway8, acts in a parallel phagocytic pathway that includes the transthyretin protein TTR-52, as well as CED-7, NRF-5 and CED-6 (refs 9, 10, 11, 12). We show that TTR-52 binds to PS exposed on the injured axon, and can restore fusion several hours after injury. We propose that PS functions as a ‘save-me’ signal for the distal fragment, allowing conserved apoptotic cell clearance molecules to function in re-establishing axonal integrity during regeneration of the nervous system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bedi, S. S. & Glanzman, D. L. Axonal rejoining inhibits injury-induced long-term changes in Aplysia sensory neurons in vitro . J. Neurosci. 21, 9667–9677 (2001)
Birse, S. C. & Bittner, G. D. Regeneration of giant axons in earthworms. Brain Res. 113, 575–581 (1976)
Deriemer, S. A., Elliott, E. J., Macagno, E. R. & Muller, K. J. Morphological evidence that regenerating axons can fuse with severed axon segments. Brain Res. 272, 157–161 (1983)
Ghosh-Roy, A., Wu, Z., Goncharov, A., Jin, Y. & Chisholm, A. D. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. J. Neurosci. 30, 3175–3183 (2010)
Hoy, R. R., Bittner, G. D. & Kennedy, D. Regeneration in crustacean motoneurons: evidence for axonal fusion. Science 156, 251–252 (1967)
Macagno, E. R., Muller, K. J. & DeRiemer, S. A. Regeneration of axons and synaptic connections by touch sensory neurons in the leech central nervous system. J. Neurosci. 5, 2510–2521 (1985)
Neumann, B., Nguyen, K. C., Hall, D. H., Ben-Yakar, A. & Hilliard, M. A. Axonal regeneration proceeds through specific axonal fusion in transected C. elegans neurons. Dev. Dyn. 240, 1365–1372 (2011)
Wang, X. et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 302, 1563–1566 (2003)
Liu, Q. A. & Hengartner, M. O. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans . Cell 93, 961–972 (1998)
Wang, X. et al. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nature Cell Biol. 12, 655–664 (2010)
Wu, Y. C. & Horvitz, H. R. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93, 951–960 (1998)
Zhang, Y., Wang, H., Kage-Nakadai, E., Mitani, S. & Wang, X. C. elegans secreted lipid-binding protein NRF-5 mediates PS appearance on phagocytes for cell corpse engulfment. Curr. Biol. 22, 1276–1284 (2012)
Pérez-Vargas, J. et al. Structural basis of eukaryotic cell–cell fusion. Cell 157, 407–419 (2014)
Zeev-Ben-Mordehai, T., Vasishtan, D., Siebert, C. A. & Grunewald, K. The full-length cell–cell fusogen EFF-1 is monomeric and upright on the membrane. Nat. Commun. 5, 3912 (2014)
Mohler, W. A. et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell 2, 355–362 (2002)
Podbilewicz, B. Cell fusion. WormBook http://dx.doi.org/10.1895/wormbook.1.52.1 (2006)
Oren-Suissa, M., Hall, D. H., Treinin, M., Shemer, G. & Podbilewicz, B. The fusogen EFF-1 controls sculpting of mechanosensory dendrites. Science 328, 1285–1288 (2010)
Fadok, V. A. et al. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148, 2207–2216 (1992)
Fadok, V. A. et al. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 405, 85–90 (2000)
Yang, H. et al. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat. Commun. http://dx.doi.org/10.1038/ncomms6717 (in the press)
Chang, B., Chen, Y., Zhao, Y. & Bruick, R. K. JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007)
Hong, X. et al. Interaction of JMJD6 with single-stranded RNA. Proc. Natl Acad. Sci. USA 107, 14568–14572 (2010)
Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009)
Gumienny, T. L. et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 (2001)
Reddien, P. W. & Horvitz, H. R. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans . Nature Cell Biol. 2, 131–136 (2000)
Wu, Y. C. & Horvitz, H. R. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392, 501–504 (1998)
Zhou, Z., Hartwieg, E. & Horvitz, H. R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans . Cell 104, 43–56 (2001)
Mapes, J. et al. CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr. Biol. 22, 1267–1275 (2012)
Hammarlund, M., Jorgensen, E. M. & Bastiani, M. J. Axons break in animals lacking β-spectrin. J. Cell Biol. 176, 269–275 (2007)
Hammarlund, M., Nix, P., Hauth, L., Jorgensen, E. M. & Bastiani, M. Axon regeneration requires a conserved MAP kinase pathway. Science 323, 802–806 (2009)
Brenner, S. The genetics of Caenorhabditis elegans . Genetics 77, 71–94 (1974)
Zou, W. et al. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 5, e1000679 (2009)
Procko, C., Lu, Y. & Shaham, S. Glia delimit shape changes of sensory neuron receptive endings in C. elegans . Development 138, 1371–1381 (2011)
Neumann, B. & Hilliard, M. A. Loss of MEC-17 leads to microtubule instability and axonal degeneration. Cell Rep. 6, 93–103 (2014)
Mitani, S., Du, H., Hall, D. H., Driscoll, M. & Chalfie, M. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans . Development 119, 773–783 (1993)
Acknowledgements
We thank L. Hammond and S. Roy for support with microscopy; R. Tweedale for comments on the manuscript; P. Bazzicalupo, D. Hall, K. Nguyen, and members of the Hilliard laboratory for discussions and comments; and E. Jorgensen, S. Shaham and X. Wang for sharing reagents. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and the International C. elegans Gene Knockout Consortium. This work was supported by NHMRC Project Grants 631634 and 1068871, NIH R01 NS060129, and ARC Future Fellowship FT110100097 to M.A.H.; NIH grants GM059083, GM079097 and GM088241 to D.X.; HFSPO Fellowship LT000762/2012 to R.G.S.; Australian Postgraduate Award to S.C.; The University of Queensland Research Scholarship to C.L.; and ARC LIEF grant LE130100078 for microscopy.
Author information
Authors and Affiliations
Contributions
B.N., S.C., and R.G.S. carried out most experiments. C.L., E.S.L., and A.N. contributed some experiments. D.X. provided reagents, designed PSR-1 experiments, and edited the paper. B.N. and M.A.H. designed and interpreted experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Analysis of proximal–distal reconnection using compound microscopy and 3D-SIM imaging.
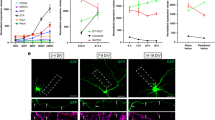
Typical examples of how reconnection was scored with compound microscopy (a, b). a, Image and schematic of a wild-type (WT) animal 24 h post-axotomy captured with a 20× objective (top panel) and two different z planes captured with a 63× objective. Despite visually appearing reconnected at 20×, the proximal regrowing axon has not made contact with the distal axon (63×); image representative of 150 animals. b, Representative images from a total of 90 animals taken with a 20× objective (top panels) and a 63× objective (bottom panels) of a WT animal 24 h post-axotomy in which reconnection has occurred between the proximal and distal axon segments. Three dimensional structured illumination microscopy (3D-SIM) was used to confirm contact regions between regrowing and distal axon fragments in WT (c) and eff-1(ok1021) mutant (d) animals. In each case that reconnection was observed with compound microscopy (63×), it was also evident with 3D-SIM imaging; n = 14 for each genotype. c, Successful reconnection in a WT animal, 24 h post-axotomy. Top panel: single-plane image acquired with a 20× objective on a compound microscope. Bottom panels: single-plane images acquired with a 63× objective on a compound microscope (left, 63×) and a SIM image of the same region (right, 63× SIM); images representative of 18 animals. d, Proximal–distal reconnection in an eff-1(ok1021) mutant animal, 24 h post-axotomy. Top panel: single-plane image acquired with a 20× objective on a compound microscope. Middle panel: single-plane images acquired with a 63× objective on a compound microscope (63×), showing two different z-planes where the regions indicated by brackets as ‘contact’ zones are in focus. Bottom panel: a SIM image of the same regions highlighting the points of contact; images representative of 14 animals. Arrowheads point to the site of axotomy, and brackets highlight zones of either potential (a) or actual reconnection (b–d). Dashed boxes represent regions magnified in the 63× images. Scale bars, 25 μm (a, b and 20× images in c, d) or 5 µm (63× images in c, d). Estimated resolution of SIM images (as per Zeiss software ZEN Black 2012): 120 nm in (c) and 110 nm in (d).
Extended Data Figure 2 EFF-1 localization in PLM neurons before and after axotomy.
Maximum projections of z-stack confocal images of an uncut axon and regrowing axons carrying the transgene Pmec-4::EFF-1::GFP; Pmec-4::mCherry in an eff-1(ok1021) mutant background. Representative images are shown for an intact axon (a), and axotomized axons 30 min post-axotomy (b), 3 h post-axotomy (c), and 8 h post-axotomy (d). a, In uncut axons, EFF-1 is present in the cell body (excluding the nucleus), posterior process and all along the axon in an irregular pattern. b, Thirty minutes after axotomy, EFF-1 forms a ‘cap’ on both the proximal (arrow) and distal (arrowhead) fragments. c, After 3 h, the proximal stump begins to swell. ‘Capping’ of the proximal (arrow) and distal (arrowhead) fragments by EFF-1 is still present. d, After 8 h, a growth cone with filopodia-like protrusions forms from the proximal stump. EFF-1 is present on the membrane of the growth cone (arrow) and filopodia-like structures (arrowheads). Scale bars, 10 µm. Asterisks highlight intestinal autofluorescence. Images representative of 15 (a), 12 (b), 9 (c), and 7 (d) animals.
Extended Data Figure 3 EFF-1 localizes to the membrane of growth cones, and functions genetically downstream from the apoptotic genes.
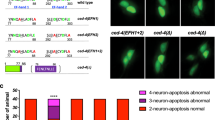
a, Maximum projection of z-stack confocal images of a regrowing axon carrying the transgene Pmec-4::EFF-1::GFP; Pmec-4::mCherry in eff-1(ok1021) mutant background, 24 h post-axotomy. EFF-1 is present on the membrane of the growth cone (line scans 1, 2 and 3) and forms a ‘cap’ at the proximal tip of the distal fragment (arrowhead). Asterisk shows the scar at the cut site. Scale bars, 10 µm. Images representative of 8 animals. b, Fluorescence profiles from the line scans shown in the left panels of panel a illustrate the presence of EFF-1::GFP on the membrane of the growth cone (1 to 3) but not on the membrane of a putative fusion site (line scan 4). au, arbitrary units. c, Successful axonal fusion in single mutant animals (white bars) compared to double mutants with eff-1(ok1021) (grey bars). Double mutants do not show a worsening of the defect compared to eff-1 single mutants. d, Cell-autonomous overexpression of EFF-1 (white bars) in wild-type (WT) animals, or in those carrying mutations in psr-1 or ttr-52, rescuing their defect. For c and d, error bars represent standard error of proportion. n values shown adjacent to each bar. P values from t-test: *P < 0.05, **P < 0.01, ***P < 0.001 compared to WT unless otherwise indicated. ns, not significant.
Extended Data Figure 4 Analysis of the localization of functional fluorophore-tagged PSR-1.
a, Maximum projection confocal images showing the co-localization of PSR-1::mCherry (top panel) with a mitochondrial marker (TOMM-20::GFP, middle panel) in a psr-1(ok714) mutant animal. PSR-1 displays strong co-localization with the mitochondria; however, there are regions where co-localization is not present, and these are highlighted by the arrow and arrowheads in the overlay image (bottom panel). Images are representative of 35 animals. Animals of the same genotype were analysed before and after axotomy (b) to determine the localization of PSR-1 compared to the mitochondrial marker during regrowth. PSR-1 accumulates to the end of the regrowing proximal axon, whereas the TOMM-20::GFP remains restricted to the mitochondria; images representative of 27 animals. Panels to the right show magnified views of the boxed regions; arrowheads point to the site of axotomy. Scale bars, 25 µm.
Extended Data Figure 5 Analysis of axonal fusion and regeneration after heat-shock treatment in wild-type and ttr-52 mutant animals, and analysis of sAnxV localization after axotomy.
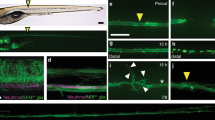
a, Animals mutant in ced-2, ced-5, ced-12, or ced-10 have normal axonal fusion similar to that of wild-type (WT) animals. b, Unsuccessful fusion in a ttr-52(sm211) animal. Regrowth has extended from the cut site (arrowhead) and contacted the distal fragment (bracketed region), but degeneration of the distal fragment has begun. Scale bar, 25 µm. Image representative of 16 animals. c, Rate of successful axonal fusion in ttr-52(sm211) animals without or with heat-shock (HS) treatment. d, The average length of regrowth in ttr-52(sm211) animals given HS after axotomy compared to those without HS treatment. e, Quantification of regrowth in ttr-52(sm211); Phsp::TTR-52::mCherry animals without HS treatment compared to those that received HS immediately after axotomy (0 h), or at 6 or 9 h post-axotomy. f, Comparison of the average length of regrowth in zdIs5 animals without HS or with HS given at different times after axotomy. g, Successful axonal fusion in zdIs5 animals without HS compared to animals given HS at different times post-axotomy. Error bars represent standard error of proportion (a, c, g) and standard error (d–f). n values shown adjacent to each bar. P values (from t-test for a, c, g; from Kolmogorov–Smirnov test for d–f): **P < 0.01, ***P < 0.001 compared to WT or no HS (black bars), unless marked otherwise on the graph. ns, not significant. h, Single plane confocal images showing the localization of the PS sensor, sAnxV::mRFP, to the axon of PLM before axotomy and at several time points after transection. The overlay image shows the 60 min time-point; filled arrowhead points to the site of axotomy, and asterisks highlight the accumulation of sAnxV::mRFP at the injury site. Scale bar, 25 μm. Images representative of 26 animals.
Extended Data Figure 6 Quantification of regeneration.
a, To determine if the loss of any of the genes analysed caused a reduction in the regenerative potential of PLM, we calculated the average length of regrowth over the first 24 h post-axotomy. Mutations in several genes that reduced axonal fusion, including psr-1, ttr-52, nrf-5 and ced-7, caused defective regrowth in PLM. However, we found no correlation between the average length of regrowth and the rate of axonal fusion. For instance, ced-10 and ced-12 mutants displayed reduced regrowth (Supplementary Table 2), but presented no defect in axonal fusion (Extended Data Fig. 5a), whereas animals that lacked eff-1 had the strongest defect in axonal fusion, but their regenerative growth was not different from that of wild-type (WT) animals. Moreover, there was no correlation between axonal fusion and the percentage of animals able to initiate any regrowth from the proximal axon after transection (b), the average number of axons that were able to regrow across the cut site at different time points after axotomy (c), or the percentage of axons presenting a growth cone at different stages of regeneration (d). Error bars represent standard error (a) or standard error of proportion (b–d). n values are displayed in Supplementary Table 2. P values (from Kolmogorov–Smirnov test for a; from t-test for b–d): *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Figure 7 Localization of CED-6, CED-7, and NRF-5 in PLM before and after axotomy.
a, Cell-autonomous rescue of the axonal fusion defect in ced-6(n1813) and ced-7(n2690) animals with expression in the mechanosensory neurons of either wild-type ced-6 genomic DNA, or ced-7 cDNA, tagged with mRFP. Error bars represent standard error of proportion; n values displayed in graph. P values from t-test: *P < 0.05. b, Maximum projection confocal images highlighting co-localization between mRFP::CED-6 and a mitochondrial marker (TOMM-20::GFP) in a ced-6(n1813) mutant background. Arrowheads point to regions where this co-localization is absent. Scale bar, 25 µm. Images representative of 12 animals. c, Single plane confocal images showing the localization of mRFP::CED-6 in PLM in a ced-6(n1813) mutant animal before axotomy (top two panels), and 4 h post-axotomy (bottom three panels). Panels to the right show magnified images of the boxed regions to highlight accumulation of CED-6 at the end of the regrowing proximal axon. Arrowheads point to the site of axotomy. Scale bars, 25 µm; images representative of 9 animals. d, CED-7::mRFP localization in single plane confocal images of PLM in ced-7(n2690) mutant animals without axotomy (top panels) and at several time points after axotomy. Different animals are shown for each time point. Filled arrowheads point to the site of axotomy; open arrowhead points to site of fusion. Scale bars, 25 µm. Images representative of 10 animals. e, Single plane confocal images of the localization of NRF-5::mCherry (expressed from a heat-shock promoter) in nrf-5 mutants to the PLM axon before axotomy and at several time points after transection in animals with genotype nrf-5(sa513); qxIs92; zdIs5. Overlay image is shown for the 60 min time-point; arrowheads point to the site of axotomy. Scale bar, 25 μm. Images representative of 29 animals. f, Quantification of the relative fluorescence levels of NRF-5::mCherry on the PLM distal (blue line) and proximal (red line) axons after axotomy compared to mock surgery conditions (black line). Error bars represent standard error; n = 29 for distal and proximal axon, n = 13 for mock surgery. P values from Kolmogorov–Smirnov test: *P < 0.05 compared to mock surgery.
Extended Data Figure 8 Mutation of unc-70 causes axonal defects and PLM-PLN fusion events that are dependent on eff-1 and dlk-1.
a, In the absence of UNC-70 the PLM mechanosensory neurons develop abnormal morphology with breaks, loop structures (arrowhead) and branching. Scale bar, 25 µm; image representative of 21 animals. b, Quantification of the penetrance of abnormal loop structures as well as axonal breakages; n value within the bar. c, To determine whether the fluorescence observed in PLN was a result of fusion with PLM, or due to a promoter-switching event, time-lapse imaging was undertaken in vivo, in unc-70(s1502) mutant animals. Single plane images captured on a spinning disk confocal are shown. Asterisk denotes PLM cell body (i); filled arrowhead indicates the site of initial GFP transfer first visible at 0 h (i); open arrowhead indicates site of eventual PLN cell body appearance, which occurs after GFP is visible in the axonal process (ii–ix). Scale bar, 50 μm. Image representative of one fusion event captured from 23 animals imaged. d, To further confirm the fusion events in unc-70(s1502) mutants, a tethered fluorophore assay was used to analyse cytoplasmic movement between PLM and PLN. Cytoplasmic GFP was expressed together with mRFP targeted to the mitochondria (TOMM-20::mRFP), both expressed under the control of the Pmec-4 promoter in the same extra-chromosomal array. In wild-type (WT) animals, both GFP and TOMM-20::mRFP are only visible in the soma and axon of the PLM neuron (no extra cell was visible). In those unc-70(s1502) mutant animals that presented PLM and PLN spontaneous neuronal fusion visible as GFP in both cells, PLM appeared as per wild-type with TOMM-20::mRFP in the soma and axon (left bar), whereas in PLN (designated ‘Extra’ in the bar) mitochondria were never observed. Animals with mutations in sem-4(n1971) and ced-3(n717) contain extra cells due to cell fate defects35; as a result, both GFP and TOMM-20::mRFP are visible in the axon and soma of both PLM and the extra cells. Error bars represent standard error of proportion; n values for cells analysed in each bar. e, Spontaneous fusion between PLM and PLN is completely abolished in dlk-1 mutants, and shows a large reduction in an eff-1 mutant background. Overexpression of EFF-1 in the PLM neuron causes a large enhancement in spontaneous fusion events, but in animals lacking DLK-1, overexpression of EFF-1 in PLM is not sufficient to induce neuronal fusion between PLM and PLN. For d and e, error bars represent standard error of proportion. n values are shown adjacent to each bar. P values from t-test: ***P < 0.001.
Extended Data Figure 9 TTR-52 and sAnxV bind to the PLM axon in unc-70 mutants.
Localization of TTR-52::mCherry to the PLM axon at the L4 stage (a) or one-day-old adult (1DOA) animal (b) of unc-70 mutant animals (genotype: unc-70(s1502); oxIs95; smIs119; zdIs5). Scale bars, 20 μm. Images representative of 16 (a) and 17 (b) animals. c, Quantification of the relative fluorescence levels of TTR-52::mCherry on the PLM axons in L4 and 1DOA wild-type (WT) animals compared to unc-70(s1502) mutants. The intensity of TTR-52::mCherry on the axon shows a small but significant increase in unc-70(s1502) mutants compared to WT animals at the L4 stage and a more pronounced enhancement in 1DOA animals; n = 15 for WT L4, n = 16 for unc-70 L4, and n = 17 for both genotypes at 1DOA stage. d, Localization of the PS sensor, sAnxV::mRFP, to the PLM axon in an unc-70(s1502) mutant animal at the L4 stage. e, Localization of sAnxV::mRFP to the PLM axon in a 1DOA unc-70(s1502) mutant animal. Scale bars, 20 μm. Images representative of 14 (d) and 15 (e) animals. f, Quantification of the relative fluorescence levels of sAnxV::mRFP on the PLM axons in L4 and 1DOA WT animals compared to unc-70(s1502) mutants. In 1DOA animals there is a large increase in the relative fluorescence in unc-70(s1502) mutants compared to WT animals; n = 15 for WT L4, n = 14 for unc-70 L4, n = 14 for WT 1DOA, and 15 for unc-70 1DOA. For c and f, error bars indicate standard error. P values from Kolmogorov–Smirnov test: *P < 0.05, ***P < 0.001. ns, not significant. au, arbitrary units.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2, a Supplementary Discussion and Supplementary References. (PDF 186 kb)
Rights and permissions
About this article
Cite this article
Neumann, B., Coakley, S., Giordano-Santini, R. et al. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature 517, 219–222 (2015). https://doi.org/10.1038/nature14102
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14102
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.