Abstract
The major facilitator superfamily glucose transporters, exemplified by human GLUT1–4, have been central to the study of solute transport. Using lipidic cubic phase crystallization and microfocus X-ray diffraction, we determined the structure of human GLUT3 in complex with d-glucose at 1.5 Å resolution in an outward-occluded conformation. The high-resolution structure allows discrimination of both α- and β-anomers of d-glucose. Two additional structures of GLUT3 bound to the exofacial inhibitor maltose were obtained at 2.6 Å in the outward-open and 2.4 Å in the outward-occluded states. In all three structures, the ligands are predominantly coordinated by polar residues from the carboxy terminal domain. Conformational transition from outward-open to outward-occluded entails a prominent local rearrangement of the extracellular part of transmembrane segment TM7. Comparison of the outward-facing GLUT3 structures with the inward-open GLUT1 provides insights into the alternating access cycle for GLUTs, whereby the C-terminal domain provides the primary substrate-binding site and the amino-terminal domain undergoes rigid-body rotation with respect to the C-terminal domain. Our studies provide an important framework for the mechanistic and kinetic understanding of GLUTs and shed light on structure-guided ligand design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nelson, D. L. & Cox, M. M. Lehninger Principles of Biochemistry (W. H. Freeman, 2008)
Hediger, M. A., Clemencon, B., Burrier, R. E. & Bruford, E. A. The ABCs of membrane transporters in health and disease (SLC series): introduction. Mol. Aspects Med. 34, 95–107 (2013)
Mueckler, M. & Thorens, B. The SLC2 (GLUT) family of membrane transporters. Mol. Aspects Med. 34, 121–138 (2013)
Manolescu, A. R., Witkowska, K., Kinnaird, A., Cessford, T. & Cheeseman, C. Facilitated hexose transporters: new perspectives on form and function. Physiology 22, 234–240 (2007)
LeFevre, P. G. Evidence of active transfer of certain non-electrolytes across the human red cell membrane. J. Gen. Physiol. 31, 505–527 (1948)
Kasahara, M. & Hinkle, P. C. Reconstitution and purification of the d-glucose transporter from human erythrocytes. J. Biol. Chem. 252, 7384–7390 (1977)
Mueckler, M. et al. Sequence and structure of a human glucose transporter. Science 229, 941–945 (1985)
Thorens, B. & Mueckler, M. Glucose transporters in the 21st century. Am. J. Physiol. Endocrinol. Metab. 298, E141–E145 (2010)
Dick, A. P., Harik, S. I., Klip, A. & Walker, D. M. Identification and characterization of the glucose transporter of the blood-brain barrier by cytochalasin B binding and immunological reactivity. Proc. Natl Acad. Sci. USA 81, 7233–7237 (1984)
Maher, F., Vannucci, S. J. & Simpson, I. A. Glucose transporter proteins in brain. FASEB J. 8, 1003–1011 (1994)
Thorens, B., Sarkar, H. K., Kaback, H. R. & Lodish, H. F. Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and β-pancreatic islet cells. Cell 55, 281–290 (1988)
Fukumoto, H. et al. Sequence, tissue distribution, and chromosomal localization of mRNA encoding a human glucose transporter-like protein. Proc. Natl Acad. Sci. USA 85, 5434–5438 (1988)
Simpson, I. A. et al. The facilitative glucose transporter GLUT3: 20 years of distinction. Am. J. Physiol. Endocrinol. Metab. 295, E242–E253 (2008)
James, D. E., Brown, R., Navarro, J. & Pilch, P. F. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein. Nature 333, 183–185 (1988)
Birnbaum, M. J. Identification of a novel gene encoding an insulin-responsive glucose transporter protein. Cell 57, 305–315 (1989)
Mueckler, M. Facilitative glucose transporters. Eur. J. Biochem. 219, 713–725 (1994)
Pascual, J. M. et al. GLUT1 deficiency and other glucose transporter diseases. Eur. J. Endocrinol. 150, 627–633 (2004)
Santer, R. et al. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi–Bickel syndrome. Nature Genet. 17, 324–326 (1997)
Simpson, I. A., Chundu, K. R., Davies-Hill, T., Honer, W. G. & Davies, P. Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer’s disease. Ann. Neurol. 35, 546–551 (1994)
Macheda, M. L., Rogers, S. & Best, J. D. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J. Cell. Physiol. 202, 654–662 (2005)
Amann, T. & Hellerbrand, C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin. Ther. Targets 13, 1411–1427 (2009)
Shim, B. Y. et al. Glucose transporter 1 (GLUT1) of anaerobic glycolysis as predictive and prognostic values in neoadjuvant chemoradiotherapy and laparoscopic surgery for locally advanced rectal cancer. Int. J. Colorectal Dis. 28, 375–383 (2013)
Ramani, P., Headford, A. & May, M. T. GLUT1 protein expression correlates with unfavourable histologic category and high risk in patients with neuroblastic tumours. Virchows Arch. 462, 203–209 (2013)
Flavahan, W. A. et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nature Neurosci. 16, 1373–1382 (2013)
Airley, R. E. & Mobasheri, A. Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: novel pathways and targets for anticancer therapeutics. Chemotherapy 53, 233–256 (2007)
Kaira, K. et al. Biological significance of 18F-FDG uptake on PET in patients with non-small-cell lung cancer. Lung Cancer 83, 197–204 (2013)
Gallamini, A., Zwarthoed, C. & Borra, A. Positron emission tomography (PET) in oncology. Cancers 6, 1821–1889 (2014)
Calvaresi, E. C. & Hergenrother, P. J. Glucose conjugation for the specific targeting and treatment of cancer. Chem. Sci. 4, 2319–2333 (2013)
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966)
Shi, Y. Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 42, 51–72 (2013)
Smirnova, I., Kasho, V. & Kaback, H. R. Lactose permease and the alternating access mechanism. Biochemistry 50, 9684–9693 (2011)
Deng, D. et al. Crystal structure of the human glucose transporter GLUT1. Nature 510, 121–125 (2014)
Sun, L. et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490, 361–366 (2012)
Quistgaard, E. M., Low, C., Moberg, P., Tresaugues, L. & Nordlund, P. Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters. Nature Struct. Mol. Biol. 20, 766–768 (2013)
Wisedchaisri, G., Park, M. S., Iadanza, M. G., Zheng, H. & Gonen, T. Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nature Commun. 5, 4521 (2014)
Lacko, L., Wittke, B. & Geck, P. The temperature dependence of the exchange transport of glucose in human erythrocytes. J. Cell. Physiol. 82, 213–218 (1973)
Chen, C. C. et al. Human erythrocyte glucose transporter: normal asymmetric orientation and function in liposomes. Proc. Natl Acad. Sci. USA 83, 2652–2656 (1986)
Rumsey, S. C. et al. Glucose transporter isoforms GLUT1 and GLUT3 transport dehydroascorbic acid. J. Biol. Chem. 272, 18982–18989 (1997)
Rumsey, S. C. et al. Dehydroascorbic acid transport by GLUT4 in Xenopus oocytes and isolated rat adipocytes. J. Biol. Chem. 275, 28246–28253 (2000)
Uldry, M., Ibberson, M., Hosokawa, M. & Thorens, B. GLUT2 is a high affinity glucosamine transporter. FEBS Lett. 524, 199–203 (2002)
Maher, F., Davies-Hill, T. M. & Simpson, I. A. Substrate specificity and kinetic parameters of GLUT3 in rat cerebellar granule neurons. Biochem. J. 315, 827–831 (1996)
Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 38, 151–159 (2013)
Carruthers, A. & Helgerson, A. L. Inhibitions of sugar transport produced by ligands binding at opposite sides of the membrane. Evidence for simultaneous occupation of the carrier by maltose and cytochalasin B. Biochemistry 30, 3907–3915 (1991)
Colville, C. A., Seatter, M. J., Jess, T. J., Gould, G. W. & Thomas, H. M. Kinetic analysis of the liver-type (GLUT2) and brain-type (GLUT3) glucose transporters in Xenopus oocytes: substrate specificities and effects of transport inhibitors. Biochem. J. 290, 701–706 (1993)
Janoshazi, A. & Solomon, A. K. Initial steps of alpha- and beta-d-glucose binding to intact red cell membrane. J. Membr. Biol. 132, 167–178 (1993)
Leitch, J. M. & Carruthers, A. α- and β-monosaccharide transport in human erythrocytes. Am. J. Physiol. Cell Physiol. 296, C151–C161 (2009)
Carruthers, A. & Melchior, D. L. Transport of alpha- and beta-d-glucose by the intact human red cell. Biochemistry 24, 4244–4250 (1985)
Barnett, J. E., Holman, G. D. & Munday, K. A. An explanation of the asymmetric binding of sugars to the human erythrocyte sugar-transport systems. Biochem. J. 135, 539–541 (1973)
DeLano, W. L. The PyMOL molecular graphics system (Schrödinger, 2002)
Smart, O. S., Goodfellow, J. M. & Wallace, B. A. The pore dimensions of gramicidin A. Biophys. J. 65, 2455–2460 (1993)
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nature Protocols 4, 706–731 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Stein, N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 (2008)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002)
Echols, N., Milburn, D. & Gerstein, M. MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 31, 478–482 (2003)
Krebs, W. G. & Gerstein, M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 28, 1665–1675 (2000)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Brünger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006)
Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 (2003)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994)
Acknowledgements
We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the facility support. The computation was completed on the “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology. This work was supported by funds from the Ministry of Science and Technology of China (2015CB910101, 2011CB910501, 2014ZX09507003006) and National Natural Science Foundation of China (projects 31130002 and 31125009). The research of N. Y. was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and an endowed professorship from Bayer Healthcare.
Author information
Authors and Affiliations
Contributions
N.Y. conceived the project. D.D., P.S. and N.Y. designed all experiments. D.D., P.S., C.Y., X.J., L.X., W.R., K.H., M.Y. and S.F. performed the experiments. D.D., P.S., C.Y., M.K. and N.Y. analysed the data and contributed to manuscript preparation. N.Y. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Chemical structures of the tested monosaccharides, glucose derivatives, and other chemicals in the competition assay.
a, b, The monosaccharides (a) and chemicals (b) that exhibit potent inhibition to the uptake of [2-3H]glucose in the proteoliposome-based counterflow assay are labelled brown. Those having moderate or no inhibition are labelled blue and green, respectively.
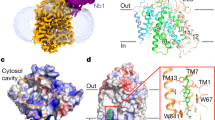
Extended Data Figure 2 Protein purification, crystallization and structural determination of GLUT3(N43T) in the presence of d-glucose.
a, A representative chromatogram of the size-exclusion chromatography purification of GLUT3. The peak positions were applied for SDS–PAGE and followed by Coomassie blue staining. b, The crystal of GLUT3 used for X-ray diffraction which led to the final structural determination at 1.5 Å resolution. c, A representative image of X-ray diffraction. The inset shows the resolution limit beyond 1.5 Å resolution. d, Crystal packing of GLUT3 in the space group of P21. Three perpendicular views are shown. Each GLUT3 is domain-coloured and shown as a ribbon. The bound glucose is shown as a black sphere and the bound lipid molecules are shown as blue sticks. e, Electron densities of three bound monoolein molecules. The 2Fo − Fc electron density maps for the bound lipid molecules are contoured at 1.0σ. f, Positions of the three bound monoolein molecules relative to the protein structure.
Extended Data Figure 3 The intracellular and extracellular gates of GLUT3.
a, The N-terminal, C-terminal and ICH domains interact with each other through an extensive network of direct and water-mediated hydrogen bonds on the intracellular side of the membrane. Water molecules are shown as red spheres. Hydrogen bonds are represented by red dashed lines. b, The polar interactions between the N-terminal and C-terminal domains on the intracellular side. An ICH domain-omitted intracellular view is shown. Note the pseudo two-fold symmetry of the overall structure and the interacting residues (inset on the right). These polar interactions partially constitute the intracellular gate of GLUT3 in the outward-facing conformation. c, Intra-domain interaction of the ICH domain. The polar interactions between helices IC3 and IC5 are shown. d, The intracellular gate of GLUT3 involves the Sugar Porter family (SP)-signature motifs. Sequence alignment of the 14 human GLUTs were performed with ClustalW65. The secondary structural elements and the SP motifs are shown on the top and bottom, respectively. The residues coloured red are the highly conserved residues that constitute the intracellular gate illustrated in panel b. e, The extracellular gate of GLUT3 in the outward-occluded state. An extracellular view is shown. f, g, The polar and hydrophobic residues mediating the lateral interactions of the N-terminal and C-terminal domains.
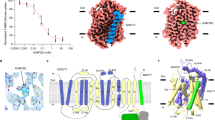
Extended Data Figure 4 d-glucose coordination by GLUT3.
a, The C-terminal domain provides the primary accommodation site for glucose in GLUT3. The α- and β-d-glucose anomers are coloured black and silver, respectively. The orange line in the right panel indicates the approximate interface between the N-terminal and C-terminal domains viewed from the extracellular side. Note that the ligand is located closer to the C-terminal domain. b, One monoolein molecule contributes to substrate coordination. The two monoolein molecules bound in the cavity of GLUT3 are coloured silver and light purple. One monoolein molecule mediates indirect hydrogen bonds between C2 and C3 hydroxyl groups of the bound glucose with the side groups of Thr28 and Gln281 of GLUT3. The effect of monoolein on glucose binding and transport has not been characterized.
Extended Data Figure 5 Attempts to obtain the structure of outward-facing GLUTs.
a, Presence of the detergent molecule β-NG helped stabilize the inward-open conformation of GLUT132. b, The glucopyranoside of β-NG and d-glucose are coordinated similarly by the inward-facing GLUT1 and the outward-facing XylE33, respectively. In both structures, the C4-OH is positioned towards the extracellular side. c, Identification of potential ligands that may stabilize GLUTs in an outward-open conformation. The indicated disaccharides, where the C4-OH of d-glucose is condensed with another hexose, were tested for their abilities to inhibit glucose transport by GLUT1 or GLUT3 in the proteoliposome-based counterflow assay. Control refers to the conditions where no competitor was added.
Extended Data Figure 6 Structure determination of GLUT3 in complex with maltose in the outward-open and outward-occluded conformations.
a, There are two GLUT3 molecules in each asymmetric unit of the outward-open structures. b, The crystal packing of the outward-open GLUT3. c, Structural superimposition of the two molecules in each asymmetric unit of the outward-open GLUT3. The two molecules exhibit nearly identical conformations except for the extracellular loop regions. The focus was on molecule (Mol) A for structural analysis and comparison in the main text. d, The crystal packing of the outward-occluded GLUT3 bound to maltose. There is only one molecule in each asymmetric unit. e, f, The 2Fo − Fc electron density maps for the bound maltose in the outward-occluded (e) and outward-open (f) structures of GLUT3, both contoured at 1σ.
Extended Data Figure 7 Maltose coordination in the outward-open and outward-occluded GLUT3.
a, Coordination of maltose in the outward-occluded GLUT3 structure. Details of the polar interactions of maltose with residues from the N-terminal and C-terminal domains are shown in the insets on the bottom and right, respectively. Hydrogen bonds are represented by brown dashed lines. b, The structure of maltose-bound outward-occluded GLUT3 is nearly identical to that of the glucose-bound GLUT3. The second glucose unit (Glc2) of maltose completely overlaps with d-glucose, which is bound to the outward-occluded GLUT3. c, The bound maltose molecules are positioned similarly in the outward-open and outward-occluded GLUT3. The two structures of GLUT3 are superimposed relative to the C-terminal domain. The outward-open GLUT3 is domain coloured and the outward-occluded structure is coloured pale purple. d, Maltose coordination in the outward-open GLUT3. Note that the coordination of Glc2 by the outward-open GLUT3 is identical to that by the outward-occluded GLUT3, while the polar residues in the N-terminal domain are not involved in the coordination of Glc1 in the outward-open structure.
Extended Data Figure 8 The molecular basis underlying the rigidity and adaptability of the N-terminal and C-terminal domains, respectively.
a, Structural feature of the N-terminal domain of GLUT3. The interior of the N-terminal domain is held through a strip of hydrogen bonds, which may provide the molecular basis for the rigidity of the N-terminal domain during the alternating access cycle. b, The C-terminal domain of GLUT3 has a hydrophobic core, which may allow the adaptability for the intra-domain shifts of the C-terminal domain during the transport cycle. c, Water distribution in the structure of glucose-bound GLUT3. The bound water molecules are shown as red spheres. Identical views are shown for the two panels except that the protein is omitted in the right panel to better illustrate the density of water molecules with respect to the N-terminal, C-terminal and ICH domains.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion on the illustration of the intra- and extracellular gates of the outward-occluded GLUT3, our endeavors for structural determination of outward-open GLUTs, and analysis of maltose-coordination by GLUT3 in the two states. Supplementary Figure 1 shows the sequence alignment of 14 human GLUTs. (PDF 1726 kb)
Conformational changes between the outward-open and outward-occluded structures of GLUT3
To better illustrate the conformational changes of the two GLUT3 structures, particularly those of TM7b and TM8, the morph was generated using the two structures of maltose-bound GLUT3 superimposed relative to their C domains. The intermediate morphs were obtained with the multiple-chain morphing script58,59 for Crystallography & NMR System (CNS)60,61. The animations were produced using PyMol. (MOV 10429 kb)
Conformational changes of the glucose-binding site during alternating access cycle
Shown here is the morph generated using the structures of GLUT1 and the glucose-bound GLUT3 superimposed to their C domains. A homology-based structural model of the inward-open GLUT3 was generated based on the structure of GLUT1 using the online SWISS-MODEL workspace62–64. The α-D-glucose is shown. For visual clarity, TM11 is omitted in the side view. (MOV 11063 kb)
Alternating access cycle of the human GLUTs
The morph was generated based on the structures of GLUT3 and GLUT1 in the three conformations as shown in Fig. 6a. A homology-based structural model of the inward-open GLUT3 was generated based on the structure of GLUT1. The modeled structure was then overlaid to the superimposed crystal structures of GLUT3 relative to their N domains. Three structures were used to generate the morph from outward-open to outward-occluded to inward-open, whereas the inward- to outward-open morph was generated based on two structures of the inward- and outward-open states. (MOV 12294 kb)
Rights and permissions
About this article
Cite this article
Deng, D., Sun, P., Yan, C. et al. Molecular basis of ligand recognition and transport by glucose transporters. Nature 526, 391–396 (2015). https://doi.org/10.1038/nature14655
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14655
This article is cited by
-
Glucose transporter 3 (GLUT3) promotes lactylation modifications by regulating lactate dehydrogenase A (LDHA) in gastric cancer
Cancer Cell International (2023)
-
Establishing mammalian GLUT kinetics and lipid composition influences in a reconstituted-liposome system
Nature Communications (2023)
-
Antidiabetic potential of Lysiphyllum strychnifolium (Craib) A. Schmitz compounds in human intestinal epithelial Caco-2 cells and molecular docking-based approaches
BMC Complementary Medicine and Therapies (2022)
-
Evolutionary balance between foldability and functionality of a glucose transporter
Nature Chemical Biology (2022)
-
Inhibition of PFKFB3 in HER2-positive gastric cancer improves sensitivity to trastuzumab by inducing tumour vessel normalisation
British Journal of Cancer (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.