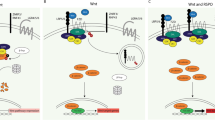
Abstract
Colorectal cancer remains a major unmet medical need, prompting large-scale genomics efforts in the field to identify molecular drivers for which targeted therapies might be developed1,2,3. We previously reported the identification of recurrent translocations in R-spondin genes present in a subset of colorectal tumours4. Here we show that targeting RSPO3 in PTPRK-RSPO3-fusion-positive human tumour xenografts inhibits tumour growth and promotes differentiation. Notably, genes expressed in the stem-cell compartment of the intestine were among those most sensitive to anti-RSPO3 treatment. This observation, combined with functional assays, suggests that a stem-cell compartment drives PTPRK-RSPO3 colorectal tumour growth and indicates that the therapeutic targeting of stem-cell properties within tumours may be a clinically relevant approach for the treatment of colorectal tumours.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
Sequence data have been deposited at the European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/home) under accession number EGAS00001001462.
References
The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012)
De Sousa E Melo, F. et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nature Med. 19, 614–618 (2013)
Sadanandam, A. et al. Reconciliation of classification systems defining molecular subtypes of colorectal cancer: interrelationships and clinical implications. Cell Cycle 13, 353–357 (2014)
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012)
Siegel, R., Desantis, C. & Jemal, A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 64, 104–117 (2014)
Kim, K. A. et al. R-spondin proteins: a novel link to beta-catenin activation. Cell Cycle 5, 23–26 (2006)
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011)
Hao, H. X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012)
Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 (2013)
Vermeulen, L. et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nature Cell Biol. 12, 468–476 (2010)
Schuijers, J. et al. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell 16, 158–170 (2015)
Jung, P. et al. Isolation and in vitro expansion of human colonic stem cells. Nature Med. 17, 1225–1227 (2011)
Kosinski, C. et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl Acad. Sci. USA 104, 15418–15423 (2007)
Dow, L. E. et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 161, 1539–1552 (2015)
Madan, B. et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene (2015)
Scholer-Dahirel, A. et al. Maintenance of adenomatous polyposis coli (APC)-mutant colorectal cancer is dependent on Wnt/beta-catenin signaling. Proc. Natl Acad. Sci. USA 108, 17135–17140 (2011)
Yan, K. S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl Acad. Sci. USA 109, 466–471 (2012)
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009)
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5 . Nature 449, 1003–1007 (2007)
Metcalfe, C., Kljavin, N. M., Ybarra, R. & de Sauvage, F. J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 14, 149–159 (2014)
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011)
O’Brien, C. A., Pollett, A., Gallinger, S. & Dick, J. E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110 (2007)
Ricci-Vitiani, L. et al. Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115 (2007)
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J. & Clarke, M. F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA 100, 3983–3988 (2003)
Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Med. 3, 730–737 (1997)
Kreso, A. & Dick, J. E. Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–291 (2014)
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015)
Wu, C. et al. RSPO2–LGR5 signaling has tumour-suppressive activity in colorectal cancer. Nature Commun. 5, 3149 (2014)
Fevr, T., Robine, S., Louvard, D. & Huelsken, J. Wnt/b-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559 (2007)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Tso, K. Y., Lee, S. D., Lo, K. W. & Yip, K. Y. Are special read alignment strategies necessary and cost-effective when handling sequencing reads from patient-derived tumor xenografts? BMC Genomics 15, 1172 (2014)
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 43, 491–498 (2011)
Saunders, C. T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012)
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001)
Fu, W. et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220 (2013)
Forbes, S. A. et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 38, D652–D657 (2010)
Acknowledgements
We would like to thank D. Jakubiak, Z. Modrusan, J. Stinson, P. Haverty and the Sequencing and Microarray laboratories for assistance with the animal studies and RNA-seq data generation.
Author information
Authors and Affiliations
Contributions
E.E.S. collected and analysed data from antibody characterization, tumour response characterization, normal intestinal response experiments, and wrote the manuscript. S.D. analysed the RNA-seq data and performed the GSEA. F.d.S.e.M. assisted in the analysis of the RNA-seq and flow cytometry data. J.T. and E.B. assisted in the design and interpretation of efficacy studies. N.K. assisted in the irradiation experiments. C.T., C.C., J.-A.H. and Y.W. generated and assisted in the characterization of the antibodies. T.B. purified proteins and antibodies. R.F. assisted in the histopathology. C.M. assisted with normal homeostasis experiments. E.W.S. analysed exome sequencing data. C.M., X.Y. and B.Y. screened samples to identify tumour models. T.P. generated RNA in situ hybridization data. F.d.S. was involved in study design, interpretation and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors are employees of Genentech, Inc. and E.E.S, S.D., J.T., N.K., C.T., X.Y., C.C., T.P., J.-A.H., T.B., R.F., E.B., C.M., E.W.S., R.L.Y., Y.W. and F.d.S. own shares in Roche.
Extended data figures and tables
Extended Data Figure 1 Characteristics of anti-RSPO2 and anti-RSPO3 antibodies.
a, Surface plasmon resonance sensograms of anti-RSPO2 (0–125 nM) binding to human and mouse R-spondins (as indicated; h, human; m, mouse). b, Surface plasmon resonance sensograms of anti-RSPO3 (0–125 nM) binding to human and mouse R-spondins (as indicated). c, Affinities of anti-RSPO2 and anti-RSPO3 binding to human and mouse R-spondins as indicated. Kd, equilibrium dissociation constant calculated as the ratio of koff/kon. Data are means ± s.d. d, Functional blocking of Wnt reporter assay stimulated by 11.6 pM human RSPO2 or 10.5 pM human RSPO3 and 10 ng ml−1 rmWNT3a (as indicated). Data represent the mean ± s.e.m. of three independent experiments.
Extended Data Figure 2 Tumour growth inhibition persists following anti-RSPO3 treatment cessation.
a, RNA-seq expression of RSPO1-4 and identification of RSPO3-fusion by fusion-specific quantitative reverse transcription PCR (qRT–PCR) in colon tumour, normal and PDX models. Expression data from normal colon and colon tumours were analysed from the publicly available data EGA accession EGAS00001000288 (ref. 4). Data are mean ± s.d. Normal n = 72; non-fusion tumour n = 67; PTPRK(e1)-RSPO3(e2) n = 4; CRCA n = 3 different tumours from the model; CRCB n = 3 different tumours from the model. RPKM, reads per kilobase per million. Expression levels greater than 0 but less than 0.5 RPKM are graphed as 0.5. b, Individual tumour measurements from CRCA treated with anti-RSPO3 (30 mg kg−1) twice a week for 4 weeks. Tumours were monitored for 30–40 days following the last dose of antibody (n = 10). c, Individual tumour measurements of CRCB data presented in Fig. 1. Anti-ragweed, n = 9; anti-RSPO3, n = 10. Tumour growth was followed for an additional 30–40 days following the last dose of antibody. Closed circles, anti-ragweed. Open circles, anti-RSPO3. Black bar indicates the time during which mice were treated with antibodies.
Extended Data Figure 3 Histopathological analysis of tumours from CRCA and CRCB.
Sections from CRCA and CRCB (representative tumour from n = 3 analysed) collected 4 days following the initiation of dosing and stained as indicated. CC3, cleaved caspase 3. Black arrows highlight extracellular MUC2 positivity, red arrows highlight intracellular MUC2 positivity. Scale bar, 100 μm.
Extended Data Figure 4 Effects of anti-RSPO3 treatment on Wnt pathway activity.
a, Heat map of a CTNNB1 gene signature29 in CRCA samples, n = 3 tumours per group. b, Heat map of a CTNNB1 gene signature29 in CRCB, n = 3 tumours per group. c, Gene set enrichment analysis of CRCA using a CTNNB1 gene signature29. RW, anti-Ragweed-treated; R3, anti-RSPO3-treated; NES, normalized enrichment score; FWER, family-wise error rate. d, Gene set enrichment analysis of CRCB using a CTNNB1 gene signature29. e, f, RNA-seq expression of a subset of Wnt target genes in CRCA (e) and CRCB (f). Data are normalized to control group. Horizontal line indicates the mean. Closed circles, anti-ragweed treated. Open circles, anti-RSPO3 treated. n = 3 tumours.
Extended Data Figure 5 Validation of gene expression changes by qRT–PCR.
a, b, Relative gene expression in CRCA and CRCB tumour samples as assessed by qRT–PCR. Closed circles, anti-ragweed-treated tumours. Open circles, anti-RSPO3-treated tumours. Horizontal line indicates the mean. n = 3 for CRCA data and n = 4 for CRCB. qRT–PCR data were validated from at least two independent experiments.
Extended Data Figure 7 APC mutant PDX models fail to respond to anti-RSPO3 treatment.
Tumour growth inhibition (left) and qRT–PCR of target genes (right) in APC mutant PDX models. a, APC(1554fs, 8fs, E941*);BRAF(V600E);PIK3CA(E545K), TP53(R248W) mutant PDX model. n = 10, anti-ragweed treated, n = 9, anti-RSPO3-treated. b, APC(757fs, E1408*);KRAS(G12D) model. Mice were treated with antibodies (30 mg kg−1) for 3–4 weeks (n = 10 animals per group). Tumour growth data were normalized to initial tumour volume upon treatment (100–150 mm3). qRT–PCR data (n = 5) were collected at 8 (a) or 4 (b) days after the initiation of dosing. Data are plotted as mean ± s.e.m. Closed circles, anti-ragweed. Open circles, anti-RSPO3. UND, undetermined Ct value.
Extended Data Figure 8 The downregulation of stem-cell marker gene expression is an early and robust response to anti-RSPO3 treatment.
a, Relative gene expression in CRCA tumour samples collected 1, 4 and 10 days following a single injection of anti-RSPO3 (30 mg kg−1). Closed circles, anti-ragweed-treated. Open circles, anti-RSPO3-treated. Horizontal line indicates the mean. n = 5 tumours per group per time point. UND, undetermined Ct value.
Extended Data Figure 9 Combined treatment of anti-RSPO2 and anti-RSPO3 reduces LGR5.
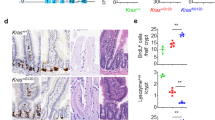
a, Quantification of Lgr5+ cells in the jejenum of mice treated with antibody (indicated on x axis) for 4 days. Lgr5–GFP-positive crypts/total crypts (n = 10 for control, n = 20 for antibody treated) were counted in three different mice. Horizontal line indicates the mean. b, Relative expression assessed by qRT–PCR from RNA isolated from the ileum of animals treated with antibody for 9 days. Data are normalized to the control antibody treated group. n = 5, horizontal line indicates the mean. c, Haematoxylin and eosin stained sections from the intestine of mice treated with indicated antibodies for 9 days. Scale bar, 100 μm.
Extended Data Figure 10 Anti-RSPO3 treatment reduces tumour-initiating cell content.
a, Percentage of CD133 (left) and CD44 (right)-positive cells as a percentage of live cells following 14 days of anti-RSPO3 treatment of CRCA. b, Percentage of CD133 (left) and CD44 (right)-positive cells as a percentage of live cells following 8 days of anti-RSPO3 treatment of CRCB. Closed circles, anti-ragweed treated. Open circles, anti-RSPO3 treated. Horizontal line indicates the mean. n = 5.
Supplementary information
Supplementary Table 1
This file contains RNAseq data of differentially expressed genes in response to anti-RSPO3 in models CRCA and CRCB. (XLSX 128 kb)
Supplementary Table 2
This file contains RNAseq data of differentially expressed genes between models CRCA and CRCB. (XLSX 656 kb)
Rights and permissions
About this article
Cite this article
Storm, E., Durinck, S., de Sousa e Melo, F. et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 529, 97–100 (2016). https://doi.org/10.1038/nature16466
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16466
This article is cited by
-
Crosstalk between colorectal CSCs and immune cells in tumorigenesis, and strategies for targeting colorectal CSCs
Experimental Hematology & Oncology (2024)
-
Clinicopathological and molecular characteristics of RSPO fusion-positive colorectal cancer
British Journal of Cancer (2022)
-
Genomic and transcriptomic analysis of Korean colorectal cancer patients
Genes & Genomics (2022)
-
Comprehensive genetic characterization of rectal cancer in a large cohort of Japanese patients: differences according to tumor location
Journal of Gastroenterology (2022)
-
NR2F2 controls malignant squamous cell carcinoma state by promoting stemness and invasion and repressing differentiation
Nature Cancer (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.