Abstract
Liver X receptors (LXRs) are transcriptional regulators of cellular and systemic cholesterol homeostasis. Under conditions of excess cholesterol, LXR activation induces the expression of several genes involved in cholesterol efflux1, facilitates cholesterol esterification by promoting fatty acid synthesis2, and inhibits cholesterol uptake by the low-density lipoprotein receptor3. The fact that sterol content is maintained in a narrow range in most cell types and in the organism as a whole suggests that extensive crosstalk between regulatory pathways must exist. However, the molecular mechanisms that integrate LXRs with other lipid metabolic pathways are incompletely understood. Here we show that ligand activation of LXRs in mouse liver not only promotes cholesterol efflux, but also simultaneously inhibits cholesterol biosynthesis. We further identify the long non-coding RNA LeXis as a mediator of this effect. Hepatic LeXis expression is robustly induced in response to a Western diet (high in fat and cholesterol) or to pharmacological LXR activation. Raising or lowering LeXis levels in the liver affects the expression of genes involved in cholesterol biosynthesis and alters the cholesterol levels in the liver and plasma. LeXis interacts with and affects the DNA interactions of RALY, a heterogeneous ribonucleoprotein that acts as a transcriptional cofactor for cholesterol biosynthetic genes in the mouse liver. These findings outline a regulatory role for a non-coding RNA in lipid metabolism and advance our understanding of the mechanisms that coordinate sterol homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tontonoz, P. Transcriptional and posttranscriptional control of cholesterol homeostasis by liver X receptors. Cold Spring Harb. Symp. Quant. Biol. 76, 129–137 (2011)
Repa, J. J. et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14, 2819–2830 (2000)
Zelcer, N., Hong, C., Boyadjian, R. & Tontonoz, P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325, 100–104 (2009)
Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997)
Zhang, Y. et al. Liver LXRα expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J. Clin. Invest. 122, 1688–1699 (2012)
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010)
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Zelcer, N. & Tontonoz, P. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Invest. 116, 607–614 (2006)
Vaisman, B. L. et al. ABCA1 overexpression leads to hyperalphalipoproteinemia and increased biliary cholesterol excretion in transgenic mice. J. Clin. Invest. 108, 303–309 (2001)
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002)
Matsuda, M. et al. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 15, 1206–1216 (2001)
Hong, C. et al. The LXR-Idol axis differentially regulates plasma LDL levels in primates and mice. Cell Metab. 20, 910–918 (2014)
Carpenter, S. et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013)
Yang, F., Zhang, H., Mei, Y. & Wu, M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell 53, 88–100 (2014)
Raal, F. J. et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet 375, 998–1006 (2010)
Gaudet, D. et al. Targeting APOC3 in the familial chylomicronemia syndrome. N. Engl. J. Med. 371, 2200–2206 (2014)
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012)
Chu, C., Quinn, J. & Chang, H. Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp . 61, 3912 (2012)
Michaud, E. J., Bultman, S. J., Stubbs, L. J. & Woychik, R. P. The embryonic lethality of homozygous lethal yellow mice (Ay/Ay) is associated with the disruption of a novel RNA-binding protein. Genes Dev. 7, 1203–1213 (1993)
Jiang, W., Guo, X. & Bhavanandan, V. P. Four distinct regions in the auxiliary domain of heterogeneous nuclear ribonucleoprotein C-related proteins. Biochim. Biophys. Acta 1399, 229–233 (1998)
Okamura, Y. et al. COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res. 43, D82–D86 (2015)
Seo, Y. K. et al. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 13, 367–375 (2011)
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, 845–858 (2015)
Auboeuf, D. et al. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 24, 442–453 (2004)
Rossi, A. et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233 (2015)
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H. & Bartel, D. P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550 (2011)
Sallam, T. et al. The macrophage LBP gene is an LXR target that promotes macrophage survival and atherosclerosis. J. Lipid Res. 55, 1120–1130 (2014)
Rong, X. et al. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 18, 685–697 (2013)
Tarling, E. J., Ahn, H. & de Aguiar Vallim, T. Q. The nuclear receptor FXR uncouples the actions of miR-33 from SREBP-2. Arterioscler. Thromb. Vasc. Biol. 35, 787–795 (2015)
Hong, C. et al. LXRα is uniquely required for maximal reverse cholesterol transport and atheroprotection in ApoE-deficient mice. J. Lipid Res. 53, 1126–1133 (2012)
Seth, P. P. et al. Short antisense oligonucleotides with novel 2′–4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 52, 10–13 (2009)
Bradley, M. N. et al. Ligand activation of LXR β reverses atherosclerosis and cellular cholesterol overload in mice lacking LXR α and apoE. J. Clin. Invest. 117, 2337–2346 (2007)
Bhatt, D. M. et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 28, 511–515 (2010)
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Carey, M. F., Peterson, C. L. & Smale, S. T. Chromatin immunoprecipitation (ChIP). Cold Spring Harb. Protoc. 2009, pdb.prot5279 (2009)
Raj, A. & Tyagi, S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472, 365–386 (2010)
Acknowledgements
We thank members of the Tontonoz, Nagy, Smale and Black laboratories and the UCLA Atherosclerosis Research Unit for technical assistance and useful discussions. This work was support by NIH grants HL030568, HL066088, DK063491, HL128822, DK102559 and HL69766; American Heart Association grant 13POST17080115; American College of Cardiology Presidential CDA; and the UCLA Cardiovascular Discovery Fund (Lauren B. Leichtman and Arthur E. Levine Investigator Award).
Author information
Authors and Affiliations
Contributions
T.S. and P.T. conceived and designed the study, guided the interpretation of the results and the preparation of the manuscript. P.T. supervised the study and provided critical suggestions. T.S. and X.W. performed most mouse experiments and data analysis. M.C.J., T.G., L.Z., J.S., C.H., T.d.A.V. participated in mouse experiments and data analysis. T.S. performed RNA-seq experiments and validated LeXis as an LXR target. A.E. and D.C. processed and analysed next-generation sequencing data. M.C.J. performed and analysed the RACE experiments. J.W. performed the mass spectrometry analysis. M.K. and R.L. provided and independently validated ASOs targeting LeXis. T.S. and P.T. drafted the manuscript. T.S., M.C.J. and P.T. edited the manuscript with input from all authors. All authors discussed the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
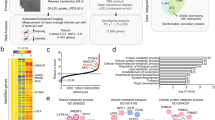
Extended Data Figure 1 Identification of LeXis as an LXR-responsive lncRNA.
a, qPCR analysis of gene expression in livers from mice gavaged with 40 mg kg−1 GW3965 for 2 days. Mice were fasted for 4 h before collection (n = 4 per group). Values are mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired two-tailed t-test). b, Volcano plot of RNA-seq results from primary hepatocytes treated for 16 h with 1 μM GW3965. c, Relative expression of selected LXR target genes identified in the RNA-seq study shown in b. Fold change represents ratio of transcript expression in GW3965 compared to DMSO treatment samples. Cut-off fold induction of 1.1 used (total 4,708 transcripts induced). d, Heat map representation of the results of transcriptional profiling (Agilent SurePrint G3 Gene Expression arrays) of primary hepatocytes treated with 1 μM GW3965 for 16 h. Data were analysed using GeneSpring software.
Extended Data Figure 2 Schematic of the LeXis gene locus and its RNA transcripts.
a, UCSC genome browser view of RNA-seq transcriptional signatures at the Abca1 and LeXis locus in mouse primary hepatocytes treated with 1 μM GW3965 for 16 h. b, Exon structure of major and minor LeXis transcripts identified by RACE, aligned for comparison to existing annotation in the indicated databases.
Extended Data Figure 3 Regulation of LeXis expression.
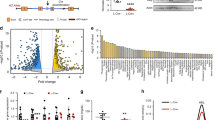
a, qPCR analysis of primary mouse hepatocytes from wild-type or double knockout (LXRα−/− and LXRβ−/−) mice treated with 1 μM GW3965 and/or 50 nM LG268. Results are representative of four independent experiments. b, LeXis expression in primary mouse hepatocytes from wild-type, LXRα−/−, LXRβ−/− or double knockout mice treated with GW3965 and LG268. Results are representative of three independent experiments. c, LeXis expression in primary hepatocytes treated with GW3965 and LG268 in the presence or absence of the protein synthesis inhibitor cycloheximide (Chx, 1 μg μl−1). Results are representative of three independent experiments. d, LeXis expression in primary hepatocytes treated with GW3965 and LG268 (50 nM) in the presence or absence of 25-hydroxycholesterol (25OH, 2.5 μM). Results are representative of three independent experiments. e, Gene expression in tissues from C57BL/6 mice gavaged with 40 mg kg−1 GW3965 for 3 days (n = 5 per group). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (unpaired two-tailed t-test). f, Relative firefly luciferase activity measured from the pgl4.10 vector or pgl4.10 with the LeXis promoter cloned upstream of luciferase. Reporters were co-transfected in HEK293 cells and treated with GW3965 for 24 h. Activity is normalized to Renilla luciferase internal control. g, Analysis of LXRα binding to the LeXis promoter in mouse liver by ChIP-qPCR. Schematic shows primer pair positions relative to the LXR-response element in the LeXis and Abca1 (positive control) promoters. Primers flanking a region of the MAP kinase I promoter served as a negative control. ChIP values are presented as percentage of input DNA (n = 4 per group). Values are mean ± s.e.m. (e, g) or mean ± s.d. (a–d). h, Prediction of coding potential using the coding-non-coding index (CNCI) software. Negative value indicates low coding potential. i, Comparison of protein coding potential using coding potential calculator (CPC) score for LeXis, the non-coding gene HOTAIR, and control protein-coding transcripts. j, In vitro translation of LeXis and luciferase control RNAs.
Extended Data Figure 4 LeXis modulates the expression of genes liked to sterol synthesis.
a, Gene expression in livers obtained after 6 days of transduction with Ad-GFP or Ad-LeXis (n = 8 per group). b, Serum alanine aminotransferase activity in chow-fed mice transduced with Ad-GFP or Ad-LeXis for 6 days (n = 8 per group). c, Gene expression in livers obtained after 6 days of transduction with Ad-GFP or Ad-LeXis (n = 8 per group). d, Unbiased pathway analysis (GeneSpring software) of the results from transcriptional profiling of livers treated with Ad-GFP or Ad-LeXis (n = 4 per group). e, Hepatic cholesterol content normalized to liver mass in wild-type mice transduced with Ad-GFP or Ad-LeXis (n = 8 per group). f, Gene expression in mouse hepatocytes treated overnight with 1 μM GW3965. Results are representative of two independent experiments. g, Gene expression in mouse hepatocytes treated overnight with Ad-GFP or Ad-LeXis for 24 h. Results are representative of two independent experiments. h, Cholesterol levels in pooled fractionated serum from Ldlr−/− mice transduced with Ad-GFP or Ad-LeXis. i, Hepatic cholesterol content normalized to liver mass in Ldlr−/− mice transduced with Ad-GFP or Ad-LeXis (n = 8 per group). j, Gene expression in livers from chow-fed wild-type or liver-specific Scap−/− mice gavaged with 40 mg kg−1 GW3965 for 2 days (n = 5 (WT Veh), 8 (WT GW), 5 (KO Veh) and 7 (KO GW)). k, Gene expression in livers from Scap−/− chow-fed mice transduced with Ad-GFP or Ad-LeXis for 6 days (n = 5 per group). Values are mean ± s.e.m. (a–c, e, i–k) or mean ± s.d. (f, g). *P < 0.05; **P < 0.01 (unpaired two-tailed t-test).
Extended Data Figure 5 Inhibition of LeXis expression alters serum cholesterol level.
a, In vitro validation of LeXis knockdown using shLeXis1 and shLeXis8 vectors. Results are representative of three independent experiments. b, Total serum cholesterol measured in C57BL/6 mice fed 2 weeks of a Western diet and transduced with adenovirus shCtrl or shLeXis8 for 6 days (n = 6–8 per group). c, Cholesterol levels in pooled fractionated serum from mice transduced with shCtrl or shLeXis adenovirus. d, Total serum cholesterol from male C57BL/6 mice fed a Western diet for 2 weeks and then transduced with control (shCtrl) or adenoviral vectors expressing shRNA targeting LeXis (shLeXis1) (n = 8 per group). e, Hepatic cholesterol content normalized to liver mass for the mice shown in d (n = 8 (shCtrl) and 7 (shLeXis1)). f, Gene expression in livers of mice fed a Western diet for 2 weeks and then transduced with shCtrl or shLeXis (n = 8 (shCtrl) and 7 (shLeXis1)). g, Total plasma cholesterol levels in chow-fed C57BL/6 mice transduced with shCtrl or shLeXis adenovirus and gavaged with 40 mg kg−1 GW3965 for 6 days (n = 8 per group). h, Gene expression in livers of chow-fed C57BL/6 mice transduced with shCtrl or shLeXis adenovirus and gavaged with 40 mg kg−1 GW3965 for 6 days (n = 8 per group). i, Serum alanine aminotransferase activity from mice in h. j, Serum alanine aminotransferase activity from mice in d. k, Gene expression in livers of mice fed a Western diet for 2 weeks and then transduced with shCtrl or shLeXis (n = 8 (shCtrl) and 7 (shLeXis1)). l, Serum alanine aminotransferase activity from C57BL/6 mice on a chow diet administered 25 mg kg−1 ASOs intraperitoneally on days 1, 4 and 7, and gavaged with 40 mg kg−1 GW3965 on days 4, 7 and 8 (n = 5 per group). m, Total serum cholesterol from C57BL/6 mice on a chow diet administered 25 mg kg−1 ASOs intraperitoneally on days 1, 3 and 5, and gavaged with 40 mg kg−1 GW3965 on days 5 and 6 (n = 8 per group). Values are mean ± s.d. (a) or mean ± s.e.m. (f, h–m). *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired two-tailed t-test (b, d–h, j) and ANOVA with multi-group comparison (m)).
Extended Data Figure 6 Generation of global LeXis−/− mice.
a, Schematic of knockout strategy. Vector construct designed to ablate entire LeXis transcript. Targeted mice were crossed with Flp−/−(also known as Hpd−/−) mice to excise the Neo cassette since it contains an active bi-directional promoter. b, c, Gene expression (n = 3 per group) and PCR genotyping strategy for LeXis−/− mice. d, Gene expression from C57BL/6 wild-type or LeXis−/− mice fed on Western diet for 3 weeks (n = 11 (WT) and 7 (LeXis−/−)). All values are mean ± s.e.m. *P < 0.05 (unpaired two-tailed t-test).
Extended Data Figure 7 Identification of RALY as a LeXis-interacting protein.
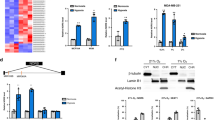
a, Complimentary biotin-labelled tiling oligonucleotides incubated with cellular extracts from liver. Probes sets designed to retrieve LeXis (Lex 1 and 2) or LacZ (LacZ 1 and 2). Percentage input of retrieved LeXis and 36B4 are shown (n = 4 per group). b, Cellular contents separated into cytoplasmic soluble (C), nuclear soluble (N) and insoluble (pellet, P) fractions were analysed by western blotting with anti-RALY and anti-histone H3 antibodies. c, Antibodies were incubated with cellular lysates from mouse hepatocytes and interaction with endogenous RALY was assessed after immunoprecipitation and western blot. d, Complexes from b were analysed for presence of LeXis or Gapdh by reverse transcription qPCR (RT–qPCR) and signals were normalized to 36B4 (n = 4 per group). e, Sequence alignment, predicted secondary structure, and 3D model of RALY are shown as reported using the Phyre2 (Protein Homology/analogueY Recognition Engine V 2.0) web portal. f, Western blot for RALY from livers transduced with adenoviral vectors expressing control shRNA (shCtrl) or Raly shRNA (shRaly) (n = pooled 4 animals per group). g, Gene expression from liver from 14-week-old chow-fed male C57BL/6 mice transduced with control (shCtrl) or shRaly (n = 8 per group). Values are mean ± s.d. (a) or mean ± s.e.m. (g).
Extended Data Figure 8 Knockdown of RALY preferentially affects pathways link to cholesterol metabolism in mouse liver.
a, b, Most significant Gene Ontology terms from microarray analysis from livers treated with shCtrl or shRaly. Analysis performed using GeneSpring and DAVID.
Extended Data Figure 9 RALY is required for LeXis mediated effects on cholesterogenesis.
a, Total serum cholesterol levels in Ldlr−/− mice transduced with shCtrl or shRaly for 6 days (n = 8 (shCtrl) and 7 (shRaly)). b, Gene expression from liver obtained from Ldlr−/− mice transduced with shCtrl or shRaly for 6 days (n = 8 (shCtrl) and 7 (shRaly)). c, Gene expression from C57BL/6 mice transduced with control (Ad-GFP) or Ad-LeXis (1.0 × 109 p.f.u.) and shCtrl or shRaly (2.0 × 109 p.f.u.) (n = 7 (ctrl/shCtrl and LeXis/shRaly) and 8 (LeXis/shCtrl and Ctrl/shRaly)). d, Recruitment of RALY in promoter regions as determined by ChIP analysis in livers transduced with control (Ad-GFP) or Ad-LeXis. Data expressed as percentage input retrieved normalized to an upstream site (region 1) (n = 3 per group). Values are mean ± s.e.m. (b, c) or mean ± s.d. (d). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (unpaired two-tailed t-test (a, b) and ANOVA with multi-group comparison (c)).
Extended Data Figure 10 Batch genome conversion between mouse and human at LeXis gene locus.
Gene expression for putative human non-coding RNA TCONS_00016452 in hepatocyte cell lines treated with 1 μM GW3965 (n = 3 per group). Values are mean ± s.d. *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired two-tailed t-test).
Supplementary information
Supplementary Information
This file contains the uncropped scans with size marker indications and primer sequences. (PDF 879 kb)
Rights and permissions
About this article
Cite this article
Sallam, T., Jones, M., Gilliland, T. et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 (2016). https://doi.org/10.1038/nature17674
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17674
This article is cited by
-
Downregulation of hepatic lncRNA Gm19619 improves gluconeogenesis and lipogenesis following vertical sleeve gastrectomy in mice
Communications Biology (2023)
-
High cholesterol intake remodels cholesterol turnover and energy homeostasis in Nile tilapia (Oreochromis niloticus)
Marine Life Science & Technology (2023)
-
Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer
Signal Transduction and Targeted Therapy (2022)
-
Long non-coding RNAs regulate fatty acid and cholesterol metabolism
Genome Instability & Disease (2022)
-
Hypothalamic long noncoding RNA AK044061 is involved in the development of dietary obesity in mice
International Journal of Obesity (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.