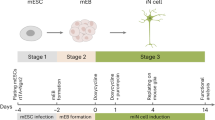
Abstract
Specification of cell identity during development depends on exposure of cells to sequences of extrinsic cues delivered at precise times and concentrations. Identification of combinations of patterning molecules that control cell fate is essential for the effective use of human pluripotent stem cells (hPSCs) for basic and translational studies. Here we describe a scalable, automated approach to systematically test the combinatorial actions of small molecules for the targeted differentiation of hPSCs. Applied to the generation of neuronal subtypes, this analysis revealed an unappreciated role for canonical Wnt signaling in specifying motor neuron diversity from hPSCs and allowed us to define rapid (14 days), efficient procedures to generate spinal and cranial motor neurons as well as spinal interneurons and sensory neurons. Our systematic approach to improving hPSC-targeted differentiation should facilitate disease modeling studies and drug screening assays.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sandoe, J. & Eggan, K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat. Neurosci. 16, 780–789 (2013).
Merkle, F.T. & Eggan, K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell 12, 656–668 (2013).
Perrimon, N., Pitsouli, C. & Shilo, B.Z. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 4, a005975 (2012).
Peljto, M. & Wichterle, H. Programming embryonic stem cells to neuronal subtypes. Curr. Opin. Neurobiol. 21, 43–51 (2011).
Guthrie, S. Patterning and axon guidance of cranial motor neurons. Nat. Rev. Neurosci. 8, 859–871 (2007).
Kanning, K.C., Kaplan, A. & Henderson, C.E. Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 33, 409–440 (2010).
Nordström, U., Maier, E., Jessell, T.M. & Edlund, T. An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 4, e252 (2006).
Amoroso, M.W. et al. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 33, 574–586 (2013).
Chen, H. et al. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 14, 796–809 (2014).
Li, X.J. et al. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 23, 215–221 (2005).
Wichterle, H., Lieberam, I., Porter, J.A. & Jessell, T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397 (2002).
Chambers, S.M. et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009).
Kirkeby, A. et al. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Reports 1, 703–714 (2012).
Mendjan, S. et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell 15, 310–325 (2014).
Bone, H.K., Nelson, A.S., Goldring, C.E., Tosh, D. & Welham, M.J. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J. Cell Sci. 124, 1992–2000 (2011).
Elkabetz, Y. et al. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 22, 152–165 (2008).
Borghese, L. et al. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells 28, 955–964 (2010).
Mazzoni, E.O. et al. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 16, 1219–1227 (2013).
Lee, S.K. & Pfaff, S.L. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38, 731–745 (2003).
Dasen, J.S., De Camilli, A., Wang, B., Tucker, P.W. & Jessell, T.M. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134, 304–316 (2008).
Miles, G.B. et al. Functional properties of motoneurons derived from mouse embryonic stem cells. J. Neurosci. 24, 7848–7858 (2004).
Pattyn, A., Hirsch, M., Goridis, C. & Brunet, J.F. Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development 127, 1349–1358 (2000).
Marklund, U. et al. Detailed expression analysis of regulatory genes in the early developing human neural tube. Stem Cells Dev. 23, 5–15 (2014).
Song, M.R. et al. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development 133, 4945–4955 (2006).
Sturgeon, K. et al. Cdx1 refines positional identity of the vertebrate hindbrain by directly repressing Mafb expression. Development 138, 65–74 (2011).
Lumsden, A. & Krumlauf, R. Patterning the vertebrate neuraxis. Science 274, 1109–1115 (1996).
Dasen, J.S. & Jessell, T.M. Hox networks and the origins of motor neuron diversity. Curr. Top. Dev. Biol. 88, 169–200 (2009).
Mazzoni, E.O. et al. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 16, 1191–1198 (2013).
Skromne, I., Thorsen, D., Hale, M., Prince, V.E. & Ho, R.K. Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development 134, 2147–2158 (2007).
Ribes, V. & Briscoe, J. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb. Perspect. Biol. 1, a002014 (2009).
Bonanomi, D. & Pfaff, S.L. Motor axon pathfinding. Cold Spring Harb. Perspect. Biol. 2, a001735 (2010).
Varela-Echavarría, A., Tucker, A., Puschel, A.W. & Guthrie, S. Motor axon subpopulations respond differentially to the chemorepellents netrin-1 and semaphorin D. Neuron 18, 193–207 (1997).
Masuda, T. et al. Netrin-1 acts as a repulsive guidance cue for sensory axonal projections toward the spinal cord. J. Neurosci. 28, 10380–10385 (2008).
Wilson, V., Olivera-Martinez, I. & Storey, K.G. Stem cells, signals and vertebrate body axis extension. Development 136, 1591–1604 (2009).
Gouti, M. et al. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 12, e1001937 (2014).
Tzouanacou, E., Wegener, A., Wymeersch, F.J., Wilson, V. & Nicolas, J.F. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell 17, 365–376 (2009).
Bialecka, M. et al. Cdx2 contributes to the expansion of the early primordial germ cell population in the mouse. Dev. Biol. 371, 227–234 (2012).
Kriks, S. et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 480, 547–551 (2011).
Xi, J. et al. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells 30, 1655–1663 (2012).
Wichterle, H. & Przedborski, S. What can pluripotent stem cells teach us about neurodegenerative diseases? Nat. Neurosci. 13, 800–804 (2010).
Sathasivam, S. Brown-Vialetto-Van Laere syndrome. Orphanet J. Rare Dis. 3, 9 (2008).
Nimchinsky, E.A. et al. Differential vulnerability of oculomotor, facial, and hypoglossal nuclei in G86R superoxide dismutase transgenic mice. J. Comp. Neurol. 416, 112–125 (2000).
Marteyn, A. et al. Mutant human embryonic stem cells reveal neurite and synapse formation defects in type 1 myotonic dystrophy. Cell Stem Cell 8, 434–444 (2011).
Di Giorgio, F.P., Boulting, G.L., Bobrowicz, S. & Eggan, K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell 3, 637–648 (2008).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Lefort, N. et al. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat. Biotechnol. 26, 1364–1366 (2008).
Nédelec, P.M., Shi, P., Amoroso, M.W., Kam, L.C. & Wichterle, H. Concentration dependent requirement for local protein synthesis in motor neuron subtype specific response to axon guidance cues. J. Neurosci. 32, 1496–1506 (2012).
Acknowledgements
I-Stem is part of the Biotherapies Institute for Rare Diseases (BIRD) supported by the Association Française contre les Myopathies (AFM-Téléthon). This project was funded by AFM, INSERM and Laboratoire d'Excellence “Labex Revive” (Investissement d'Avenir; ANR-10-LABX-73). S.N. is currently supported by the “Labex Revive” and was previously supported by a fellowship from the Genopole. N.S.-M. was supported by the program INGESTEM (investissement d'avenir ANR-11-INBS-009-01). R.A.P. and V.C. are supported by ATIP-Avenir, Agence Nationale de la Recherche (ANR-12-BSV4-0021-01, ANR-13-JSV4-0002-01) and the Ville de Paris. We thank C. Varela and N. Lefort (I-STEM) for karyotyping the cell lines and L. Aubry (I-STEM) for providing the IMR90 hiPSC line. We thank H. Wichterle, F. Giudicelli, V. Ribes, E.O. Mazzoni and A. Rebsam for helpful discussion and critical review of the manuscript. We thank J.-F. Brunet for providing the Phox2b antibody, E.R. Lowry and H. Wichterle (Columbia University) for testing the sMN differentiation and for the Hb9∷GFP-Neomycin plasmid. We thank D. Furling (Institute of Myology) for providing human primary myoblasts.
Author information
Authors and Affiliations
Contributions
Y.M., J.C., S.N., R.A.P. (electrophysiology) and N.S.-M. (real-time PCR) performed experiments. Y.M., J.C., C.M., R.A.P. and S.N. analyzed data. M.P., C.M., S.N. designed the project. S.N. and C.M. initiated the project, supervised and organized experiments. M.P. and V.C. provided resources. S.N. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Tables 1 and 2 (PDF 59363 kb)
Rights and permissions
About this article
Cite this article
Maury, Y., Côme, J., Piskorowski, R. et al. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol 33, 89–96 (2015). https://doi.org/10.1038/nbt.3049
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3049
This article is cited by
-
Generation of human iPSC-derived phrenic-like motor neurons to model respiratory motor neuron degeneration in ALS
Communications Biology (2024)
-
FUS-ALS hiPSC-derived astrocytes impair human motor units through both gain-of-toxicity and loss-of-support mechanisms
Molecular Neurodegeneration (2023)
-
Viral vector gene delivery of the novel chaperone protein SRCP1 to modify insoluble protein in in vitro and in vivo models of ALS
Gene Therapy (2023)
-
Rapid responses of human pluripotent stem cells to cyclic mechanical strains applied to integrin by acoustic tweezing cytometry
Scientific Reports (2023)
-
Self-organizing models of human trunk organogenesis recapitulate spinal cord and spine co-morphogenesis
Nature Biotechnology (2023)