Abstract
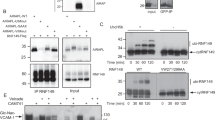
A quality-control system surveys the lumen of the endoplasmic reticulum for terminally misfolded proteins. Polypeptides singled out by this system are ultimately degraded by the cytosolic ubiquitin-proteasome pathway. Key components of both the endoplasmic reticulum quality-control system and the degradation machinery have been identified, but a connection between the two systems has remained elusive. Here, we report an association between the endoplasmic reticulum quality-control lectin Yos9p and Hrd3p, a component of the ubiquitin-proteasome system that links these pathways. We identify designated regions in the luminal domain of Hrd3p that interact with Yos9p and the ubiquitin ligase Hrd1p. Binding of misfolded proteins occurs through Hrd3p, suggesting that Hrd3p recognises proteins that deviate from their native conformation, whereas Yos9p ensures that only terminally misfolded polypeptides are degraded.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hebert, D. N., Garman, S. C. & Molinari, M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 15, 364–370 (2005).
Hitt, R. & Wolf, D. H. DER7, encoding α-glucosidase I is essential for degradation of malfolded glycoproteins of the endoplasmic reticulum. FEMS Yeast Res. 4, 815–820 (2004).
Bhamidipati, A., Denic, V., Quan, E. M. & Weissman, J. S. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell 19, 741–751 (2005).
Friedmann, E., Salzberg, Y., Weinberger, A., Shaltiel, S. & Gerst, J. E. YOS9, the putative yeast homolog of a gene amplified in osteosarcomas, is involved in the endoplasmic reticulum (ER)-Golgi transport of GPI-anchored proteins. J. Biol. Chem. 277, 35274–35281 (2002).
Jakob, C. A. et al. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2, 423–430 (2001).
Kim, W., Spear, E. D. & Ng, D. T. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell 19, 753–764 (2005).
Nakatsukasa, K., Nishikawa, S., Hosokawa, N., Nagata, K. & Endo, T. Mnl1p, an α -mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J. Biol. Chem. 276, 8635–8638 (2001).
Szathmary, R., Bielmann, R., Nita-Lazar, M., Burda, P. & Jakob, C. A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell 19, 765–775 (2005).
Meusser, B., Hirsch, C., Jarosch, E. & Sommer, T. ERAD: the long road to destruction. Nature Cell Biol. 7, 766–772 (2005).
Bays, N. W., Gardner, R. G., Seelig, L. P., Joazeiro, C. A. & Hampton, R. Y. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol. 3, 24–29 (2001).
Deak, P. M. & Wolf, D. H. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 276, 10663–10669 (2001).
Gardner, R. G. et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151, 69–82 (2000).
Plemper, R. K. et al. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell. Sci. 112, 4123–34 (1999).
Gauss, R., Sommer, T. & Jarosch, E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 25, 1827–1835 (2006).
Ponting, C. P. Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem J. 351, 527–535 (2000).
Pelham, H. R. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15, 483–486 (1990).
Kostova, Z. & Wolf, D. H. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J. Cell Sci. 118, 1485–1492 (2005).
Spear, E. D. & Ng, D. T. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J. Cell Biol. 169, 73–82 (2005).
Helenius, A. & Aebi, M. Intracellular functions of N-linked glycans. Science 291, 2364–2369 (2001).
Gemmill, T. R. & Trimble, R. B. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta. 1426, 227–237 (1999).
Ausubel, F. M. (ed.) Current Protocols in Molecular Biology (John Wiley & Sons, Inc., New York. 1993–2006).
Gauss, R., Trautwein, M., Sommer, T. & Spang, A. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast 22, 1–12 (2005).
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Knop, M. et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972 (1999).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Gueldener, U., Heinisch, J., Koehler, G. J., Voss, D. & Hegemann, J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30, e23 (2002).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Meusser, B. & Sommer, T. Vpu-mediated degradation of CD4 reconstituted in yeast reveals mechanistic differences to cellular ER-associated protein degradation. Mol. Cell 14, 247–258 (2004).
Acknowledgements
We thank C. Volkwein and A. Wittstruck for excellent technical assistance and D. H. Wolf for providing plasmids and reagents. Part of this work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) and the EU Network of Excellence RUBICON. R.G. thanks the Boehringer Ingelheim Fonds for support.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1, S2 and S3 (PDF 143 kb)
Rights and permissions
About this article
Cite this article
Gauss, R., Jarosch, E., Sommer, T. et al. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol 8, 849–854 (2006). https://doi.org/10.1038/ncb1445
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1445
This article is cited by
-
Direct observation of autoubiquitination for an integral membrane ubiquitin ligase in ERAD
Nature Communications (2024)
-
Mechanisms of substrate processing during ER-associated protein degradation
Nature Reviews Molecular Cell Biology (2023)
-
Pathway engineering facilitates efficient protein expression in Pichia pastoris
Applied Microbiology and Biotechnology (2022)
-
Hrd1 forms the retrotranslocation pore regulated by auto-ubiquitination and binding of misfolded proteins
Nature Cell Biology (2020)
-
Role of the unfolded protein response in determining the fate of tumor cells and the promise of multi-targeted therapies
Cell Stress and Chaperones (2018)