Abstract
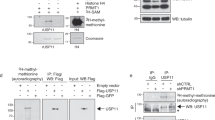
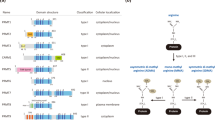
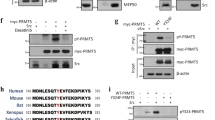
Activation of the p53 tumour suppressor protein in response to DNA damage leads to apoptosis or cell-cycle arrest. Enzymatic modifications are widely believed to affect and regulate p53 activity. We describe here a level of post-translational control that has an important functional consequence on the p53 response. We show that the protein arginine methyltransferase (PRMT) 5, as a co-factor in a DNA damage responsive co-activator complex that interacts with p53, is responsible for methylating p53. Arginine methylation is regulated during the p53 response and affects the target gene specificity of p53. Furthermore, PRMT5 depletion triggers p53-dependent apoptosis. Thus, methylation on arginine residues is an underlying mechanism of control during the p53 response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Levine, A. J. (p53, the cellular gatekeeper for growth and division. Cell 88, 323–331 1997).
Vousden, K. H. & Lu, X. Live or let die: the cell's response to p53. Nature Rev. Cancer 2, 594–604 (2002).
Levine, A. J., Hu, W. & Feng, Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 13, 1027–1036 (2006).
Coutts, A. S. & La Thangue, N. B. The p53 response: emerging levels of co-factor complexity. Biochem. Biophys. Res. Commun. 331, 778–785 (2005).
Shikama, N. et al. A novel cofactor for p300 that regulates the p53 response. Mol. Cell 4, 365–376 (1999).
Coutts, A. S., Boulahbel, H., Graham, A. & La Thangue, N. B. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 8, 84–90 (2007).
Demonacos, C., Krstic-Demonacos, M. & La Thangue, N. B. A TPR motif cofactor contributes to p300 activity in the p53 response. Mol. Cell 8, 71–84 (2001).
Demonacos, C., Krstic-Demonacos, M., Smith, L., Xu, D., O'Connor, D. P., Jansson, M. & La Thangue, N. B. A new effector pathway links ATM kinase with the DNA damage response. Nature Cell Biol. 6, 968–976 (2004).
Bedford, M. T. & Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 (2005).
Branscombe, T. L., Frankel, A., Lee, J. H., Cook, J. R., Yang, Z., Pestka, S. & Clarke, S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276, 32971–32976 (2001).
Chene, P. (2001). The role of tetramerization in p53 function. Oncogene 20, 2611–7.
Stommel, J. M. et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18, 1660–1672 (1999).
Meister, G. et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11, 1990–1994 (2001).
Pal, S., Vishwanath, S. N., Erdjument-Bromage, H., Tempst, P. & Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell Biol. 24, 9630–9645 (2004).
Rho, J. et al. PRMT5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J. Biol. Chem. 276, 11393–401 (2001).
Bunz, F. et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 (1998).
Meek, D. W. The p53 response to DNA damage. DNA Repair 3, 1049–1056 (2004).
Oda, E. et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053–1058 (2000).
Vousden, K. H. Apoptosis. p53 and PUMA: a deadly duo. Science 309, 1685–1686 (2005).
Fortin, A. et al. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J. Cell Biol. 155, 207–216 (2001).
Kastan, M. B. et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71, 587–597 (1992).
Oda, K. et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102, 849–862 (2000).
Nishi, K. et al. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269, 6320–6324 (1994).
Qian, H., Wang, T., Naumovski, L., Lopez, C. D. & Brachmann, R. K. Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene 21, 7901–7911 (2002).
Zhao, R. et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14, 981–993 (2000).
Hollstein, M. et al. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 22, 3551–3555 (1994).
Davison, T. S., Yin, P., Nie, E., Kay, C. & Arrowsmith, C. H. Characterization of the oligomerization defects of two p53 mutants found in families with Li-Fraumeni and Li-Fraumeni-like syndrome. Oncogene 17, 651–656 (1998).
Lomax, M. E., Barnes, D. M., Hupp, T. R., Picksley, S. M. & Camplejohn, R. S. Characterization of p53 oligomerization domain mutations isolated from Li-Fraumeni and Li-Fraumeni like family members. Oncogene 17, 643–649 (1998).
Kawaguchi, T. et al. The relationship among p53 oligomer formation, structure and transcriptional activity using a comprehensive missense mutation library. Oncogene 24, 6976–6981 (2005).
Stevens, C., Smith, L. & La Thangue, N. B. Chk2 activates E2F-1 in response to DNA damage. Nature Cell Biol. 5, 401–409 (2003).
Acknowledgements
We thank the MRC, Cancer Research UK, EU, LRF and AICR for supporting this work, and Rosemary Williams for assistance in preparing the manuscript.
Author information
Authors and Affiliations
Contributions
M.J., S.T.D., E.C. and S.S. designed, performed and analysed the experiments; M.E. and B.K. performed and interpreted the mass spectrometry; N.B.L.T. analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 782 kb)
Rights and permissions
About this article
Cite this article
Jansson, M., Durant, S., Cho, EC. et al. Arginine methylation regulates the p53 response. Nat Cell Biol 10, 1431–1439 (2008). https://doi.org/10.1038/ncb1802
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1802
This article is cited by
-
Regulated secretion of mutant p53 negatively affects T lymphocytes in the tumor microenvironment
Oncogene (2024)
-
PRMT5 inhibition shows in vitro efficacy against H3K27M-altered diffuse midline glioma, but does not extend survival in vivo
Scientific Reports (2024)
-
Exploiting PRMT5 as a target for combination therapy in mantle cell lymphoma characterized by frequent ATM and TP53 mutations
Blood Cancer Journal (2023)
-
PRMT5 is a therapeutic target in choroidal neovascularization
Scientific Reports (2023)
-
Autophagy dictates sensitivity to PRMT5 inhibitor in breast cancer
Scientific Reports (2023)