Abstract
Epithelial–mesenchymal transition (EMT) is associated with characteristics of breast cancer stem cells, including chemoresistance and radioresistance. However, it is unclear whether EMT itself or specific EMT regulators play causal roles in these properties. Here we identify an EMT-inducing transcription factor, zinc finger E-box binding homeobox 1 (ZEB1), as a regulator of radiosensitivity and DNA damage response. Radioresistant subpopulations of breast cancer cells derived from ionizing radiation exhibit hyperactivation of the kinase ATM and upregulation of ZEB1, and the latter promotes tumour cell radioresistance in vitro and in vivo. Mechanistically, ATM phosphorylates and stabilizes ZEB1 in response to DNA damage, ZEB1 in turn directly interacts with USP7 and enhances its ability to deubiquitylate and stabilize CHK1, thereby promoting homologous recombination-dependent DNA repair and resistance to radiation. These findings identify ZEB1 as an ATM substrate linking ATM to CHK1 and the mechanism underlying the association between EMT and radioresistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Bedford, J. S. Sublethal damage, potentially lethal damage, and chromosomal aberrations in mammalian cells exposed to ionizing radiations. Int. J. Radiat. Oncol. Biol. Phys. 21, 1457–1469 (1991).
Frankenberg-Schwager, M., Frankenberg, D., Blocher, D. & Adamczyk, C. Effect of dose rate on the induction of DNA double-strand breaks in eucaryotic cells. Radiat. Res. 87, 710–717 (1981).
Buchholz, T. A. Radiation therapy for early-stage breast cancer after breast-conserving surgery. N. Engl. J. Med. 360, 63–70 (2009).
Jameel, J. K., Rao, V. S., Cawkwell, L. & Drew, P. J. Radioresistance in carcinoma of the breast. Breast 13, 452–460 (2004).
Zhou, B. B. & Elledge, S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000).
Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K. & Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 (2004).
Parrilla-Castellar, E. R., Arlander, S. J. & Karnitz, L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 3, 1009–1014 (2004).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Reinhardt, H. C. & Yaffe, M. B. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 21, 245–255 (2009).
Wang, B., Matsuoka, S., Carpenter, P. B. & Elledge, S. J. 53BP1, a mediator of the DNA damage checkpoint. Science 298, 1435–1438 (2002).
Stewart, G. S., Wang, B., Bignell, C. R., Taylor, A. M. & Elledge, S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 (2003).
Stucki, M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 (2005).
Bao, S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006).
Baumann, M., Krause, M. & Hill, R. Exploring the role of cancer stem cells in radioresistance. Nat. Rev. Cancer 8, 545–554 (2008).
Mani, S. A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Yang, J. & Weinberg, R. A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 (2008).
Thiery, J. P., Acloque, H., Huang, R. Y. & Nieto, M. A. Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009).
Elenbaas, B. et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 15, 50–65 (2001).
Chang, C. J. et al. p53 regulates epithelial–mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 13, 317–323 (2011).
Kim, T. et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 208, 875–883 (2011).
Banath, J. P., Macphail, S. H. & Olive, P. L. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 64, 7144–7149 (2004).
Olive, P. L. & Banath, J. P. Phosphorylation of histone H2AX as a measure of radiosensitivity. Int. J. Radiat. Oncol. Biol. Phys. 58, 331–335 (2004).
Taneja, N. et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J. Biol. Chem. 279, 2273–2280 (2004).
Bauer, E. et al. The distribution of the tail moments in single cell gel electrophoresis (comet assay) obeys a chi-square (χ2) not a gaussian distribution. Mutat. Res. 398, 101–110 (1998).
Sung, P. & Klein, H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7, 739–750 (2006).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999).
Weinstock, D. M., Nakanishi, K., Helgadottir, H. R. & Jasin, M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 409, 524–540 (2006).
Smith, J., Tho, L. M., Xu, N. & Gillespie, D. A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108, 73–112 (2010).
Bartek, J. & Lukas, J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3, 421–429 (2003).
Sorensen, C. S. et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 7, 195–201 (2005).
Ma, C. X., Janetka, J. W. & Piwnica-Worms, H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol. Med. 17, 88–96 (2011).
Lukas, C. et al. DNA damage-activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res. 61, 4990–4993 (2001).
Collis, S. J. et al. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat. Cell Biol. 9, 391–401 (2007).
Leung-Pineda, V., Huh, J. & Piwnica-Worms, H. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 69, 2630–2637 (2009).
Zhang, Y. W. et al. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell 19, 607–618 (2005).
Furusawa, T., Moribe, H., Kondoh, H. & Higashi, Y. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor deltaEF1. Mol. Cell Biol. 19, 8581–8590 (1999).
Postigo, A. A. & Dean, D. C. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl Acad. Sci. USA 96, 6683–6688 (1999).
Byles, V. et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene 31, 4619–4629 (2012).
Li, M. et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 (2002).
Cummins, J. M. et al. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 428, 486 (2004).
Li, M., Brooks, C. L., Kon, N. & Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886 (2004).
Qing, P., Han, L., Bin, L., Yan, L. & Ping, W. X. USP7 regulates the stability and function of HLTF through deubiquitination. J. Cell Biochem. 112, 3856–3862 (2011).
Song, M. S. et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 (2008).
Faustrup, H., Bekker-Jensen, S., Bartek, J., Lukas, J. & Mailand, N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J. Cell Biol. 184, 13–19 (2009).
Savitsky, K. et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268, 1749–1753 (1995).
Ahn, J. Y., Schwarz, J. K., Piwnica-Worms, H. & Canman, C. E. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60, 5934–5936 (2000).
Cortez, D., Wang, Y., Qin, J. & Elledge, S. J. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286, 1162–1166 (1999).
Canman, C. E. et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 (1998).
Banin, S. et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677 (1998).
Toledo, L. I. et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 18, 721–727 (2011).
Chen, D. et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med. 18, 1511–1517 (2012).
Brabletz, T., Lyden, D., Steeg, P. S. & Werb, Z. Roadblocks to translational advances on metastasis research. Nat. Med. 19, 1104–1109 (2013).
Wang, Y. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005).
Horsman, M. R. et al. Tumor radiosensitizers—current status of development of various approaches: report of an International Atomic Energy Agency meeting. Int. J. Radiat. Oncol. Biol. Phys. 64, 551–561 (2006).
Singh, A. & Settleman, J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751 (2010).
Bartek, J. & Lukas, J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13, 738–747 (2001).
Graham, T. R. et al. Reciprocal regulation of ZEB1 and AR in triple negative breast cancer cells. Breast Cancer Res. Treat. 123, 139–147 (2010).
Karihtala, P. et al. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res. Treat. 138, 81–90 (2013).
Kenney, P. A. et al. Novel ZEB1 expression in bladder tumorigenesis. BJU Int. 107, 656–663 (2011).
Spoelstra, N. S. et al. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 66, 3893–3902 (2006).
Garrett, M. D. & Collins, I. Anticancer therapy with checkpoint inhibitors: what, where and when? Trends Pharmacol. Sci. 32, 308–316 (2011).
Liu, Y., El-Naggar, S., Darling, D. S., Higashi, Y. & Dean, D. C. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 135, 579–588 (2008).
Stewart, S. A. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003).
Yuan, J., Luo, K., Zhang, L., Cheville, J. C. & Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140, 384–396 (2010).
Richardson, C., Moynahan, M. E. & Jasin, M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12, 3831–3842 (1998).
Wang, L. et al. MK-4827, a PARP-1/-2 inhibitor, strongly enhances response of human lung and breast cancer xenografts to radiation. Invest. New Drugs 30, 2113–2120 (2012).
Acknowledgements
We thank the shRNA and ORFeome Core at MD Anderson Cancer Center and Z. Gong, A. Lin, J. Wang, W. Wang, G. Wan and X. Lu for reagents and technical assistance. We thank A. Postigo, H-L. Piao and J. Kim for discussion. This work is supported by the NIH grants R00CA138572, R01CA166051 and R01CA181029 (to L.M.) and a CPRIT Scholar Award R1004 (to L.M.). L.M. is an R. Lee Clark Fellow of The University of Texas MD Anderson Cancer Center. B.G.D. and W.A.W. are supported by a Komen Foundation Grant KG101478. Y.H. is supported in part by NIH U54CA151668. We wish to dedicate this work to the memory of K. Kian Ang.
Author information
Authors and Affiliations
Contributions
P.Z. and L.M. conceived and designed the project. P.Z. performed and analysed most of the experiments. Y.W. and M-C.H. performed studies on tissue microarrays of human patient samples. L.W. and K.K.A. performed tumour radiosensitivity studies. B.G.D. and W.A.W. established the radioresistant subline. Y.Y. and H.L. performed computational data analysis. J.Z. and D.C. made some constructs. J.Y. and J.C. provided DR-GFP-expressing U2OS cells and performed tandem-affinity purification and mass spectrometry analysis. M.W. maintained mice. Y.S. maintained shRNA and ORF clones. Y.L. and D.C.D. provided Zeb1-deficient MEFs. Y.H. contributed to discussion and revision of the manuscript. P.Z. and L.M. wrote the manuscript with input from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
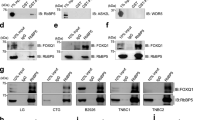
Supplementary Figure 3 Induction of EMT by Snail, Twist or ZEB1.
(a) Phase contrast images of HMLE cells transduced with Snail, Twist or ZEB1. Scale bar, 50 μm. (b) Images of clonogenic assays of HMLE cells transduced with Snail, Twist or ZEB1. (c) Immunoblotting of ZEB1 and GAPDH in mock-infected HMLE cells or HMLE cells transduced with ZEB1 alone or in combination with transfection of ZEB1 siRNA. C: control (non-irradiated); SF: survival fraction collected 3 weeks after 6-Gy irradiation.
Supplementary Figure 4 ZEB1, SNAI1 and TWIST1 mRNA levels are not substantially increased in SUM159-P2 cells.
(a) qPCR of ZEB1, SNAI1 and TWIST1 in SUM159-P0 and SUM159-P2 cells. n = 3 samples per group. (b) Clonogenic survival assays of U2OS cells transfected with ZEB1 siRNA. n = 3 wells per group. Inset: immunoblotting of ZEB1 and GAPDH. Data in a and b are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Students t-test. The experiments were repeated 3 times. The source data can be found in Supplementary Table 3. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 5 Effect of ZEB1 on CHK1, radiosensitivity and the G2 checkpoint.
(a) Immunoblotting of ZEB1, γH2AX, H2AX and GAPDH in ZEB1 shRNA-transduced SUM159-P2 cells with or without ectopic expression of an RNAi-resistant ZEB1 mutant (ZEB1-RE), at the indicated time points after 6 Gy IR. (b) Clonogenic survival assays of ZEB1 shRNA-transduced SUM159-P2 cells with or without ectopic expression of an RNAi-resistant mutant (ZEB1-RE). n = 3 wells per group. (c) Percentage of the G2/M population. SUM159-P2 cells were transduced with ZEB1 shRNA, treated with 6-Gy IR and analysed by flow cytometry. n = 3 wells per group. Data in b and c are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Students t-test. The experiments were repeated 3 times. The source data can be found in Supplementary Table 3. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 6 ZEB1 specifically regulates the protein stability of the USP7 target CHK1.
(a) qPCR of CHK1 in SUM159-P2 cells transduced with ZEB1 shRNA. n = 3 samples per group. (b) Quantification of CHK1 protein levels (normalized to GAPDH) in Fig. 5f. (c) Quantification of CHK1 protein levels (normalized to GAPDH) in Fig. 5g. (d) SUM159-P2 cells were transfected with the scramble control or USP7 siRNA (si-USP7) and then treated with 50 μg ml−1 cycloheximide (CHX). Cells were harvested at different time points as indicated and then immunoblotted with antibodies to ZEB1, USP7 and GAPDH. (e) Quantification of CHK1 protein levels (normalized to GAPDH) in Fig. 5h. (f) SUM159-P2 cells were treated with 50 μg ml−1 cycloheximide (CHX), harvested at different time points as indicated and then immunoblotted with antibodies to Claspin, ZEB1 and GAPDH. (g) 293T cells were transfected with SFB–USP7 alone or in combination with ZEB1, followed by pull-down with streptavidin-sepharose beads (s-s beads) and immunoblotting with the antibody to Claspin. Data in a are the mean of biological replicates from a representative experiment, and error bars indicate s.e.m. Statistical significance was determined by a two-tailed, unpaired Students t-test. The experiments were repeated 3 times. The source data can be found in Supplementary Table 3. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary Figure 7 ATR does not regulate ZEB1.
(a) 293T cells were transfected with SFB–ZEB1, followed by pull-down with streptavidin-sepharose beads (s-s beads) and immunoblotting with antibodies to ATR and ATM. (b) SUM159-P2 cells were transfected with ATR siRNA and treated with IR. Lysates were immunoblotted with antibodies to p-ATR, ATR, ZEB1 and GAPDH. (c) SUM159-P2 cells were pretreated with ETP-46464 and treated with IR. Lysates were immunoblotted with antibodies to ZEB1, p-CHK1, CHK1, p-CHK2, CHK2 and GAPDH.
Supplementary Figure 8 ATM-dependent phosphorylation of ZEB1 at S585 is critical for radiation-induced stabilization of ZEB1 but not the interaction between ZEB1 and USP7.
(a) 293T cells were transfected with SFB–ZEB1 (wild-type, S585A or S585D), followed by pull-down with streptavidin-sepharose beads (s-s beads) and immunoblotting with the USP7 antibody. (b,c) Quantification of ZEB1 proteins levels (normalized to co-transfected GFP) in Fig. 7i. Uncropped images of blots are shown in Supplementary Fig. 7.
Supplementary information
Supplementary Information
Supplementary Information (PDF 8268 kb)
Supplementary Table 1
Supplementary Information (XLS 114 kb)
Supplementary Table 2
Supplementary Information (XLS 30 kb)
Supplementary Table 3
Supplementary Information (XLS 50 kb)
Rights and permissions
About this article
Cite this article
Zhang, P., Wei, Y., Wang, L. et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol 16, 864–875 (2014). https://doi.org/10.1038/ncb3013
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3013
This article is cited by
-
MicroRNA-561-3p indirectly regulates the PD-L1 expression by targeting ZEB1, HIF1A, and MYC genes in breast cancer
Scientific Reports (2024)
-
Mesenchymal–epithelial transition and AXL inhibitor TP-0903 sensitise triple-negative breast cancer cells to the antimalarial compound, artesunate
Scientific Reports (2024)
-
Interaction of TAGLN and USP1 promotes ZEB1 ubiquitination degradation in UV-induced skin photoaging
Cell & Bioscience (2023)
-
ZHX2 deficiency enriches hybrid MET cells through regulating E-cadherin expression
Cell Death & Disease (2023)
-
Nuclear isoform of RAPH1 interacts with FOXQ1 to promote aggressiveness and radioresistance in breast cancer
Cell Death & Disease (2023)