Abstract
A fundamental question regarding autophagosome formation is how the shape of the double-membrane autophagosomal vesicle is generated. Here we show that in mammalian cells assembly of an actin scaffold inside the isolation membrane (the autophagosomal precursor) is essential for autophagosomal membrane shaping. Actin filaments are depolymerized shortly after starvation and actin is assembled into a network within the isolation membrane. When formation of actin puncta is disrupted by an actin polymerization inhibitor or by knocking down the actin-capping protein CapZβ, isolation membranes and omegasomes collapse into mixed-membrane bundles. Formation of actin puncta is PtdIns(3)P dependent, and inhibition of PtdIns(3)P formation by treating cells with the PI(3)K inhibitor 3-MA, or by knocking down Beclin-1, abolishes the formation of actin puncta. Binding of CapZ to PtdIns(3)P, which is enriched in omegasomes, stimulates actin polymerization. Our findings illuminate the mechanism underlying autophagosomal membrane shaping and provide key insights into how autophagosomes are formed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Klionsky, D. J. & Emr, S. D. Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 (2000).
Klionsky, D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7–18 (2005).
Lum, J. J., DeBerardinis, R. J. & Thompson, C. B. Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 6, 439–448 (2005).
Mizushima, N., Ohsumi, Y. & Yoshimori, T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27, 421–429 (2002).
Axe, E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008).
Hayashi-Nishino, M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 (2009).
Roberts, R. & Ktistakis, N. T. Omegasomes: PI3P platforms that manufacture autophagosomes. Essays Biochem. 55, 17–27 (2013).
Seglen, P. O., Gordon, P. B. & Holen, I. Non-selective autophagy. Semin. Cell Biol. 1, 441–448 (1990).
Mizushima, N. Autophagy: process and function. Genes Dev. 21, 2861–2873 (2007).
Kirchhausen, T., Owen, D. & Harrison, S. C. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6, a016725 (2014).
Shibutani, S. T. & Yoshimori, T. A current perspective of autophagosome biogenesis. Cell Res. 24, 58–68 (2014).
Chhabra, E. S. & Higgs, H. N. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110–1121 (2007).
Pollard, T. D., Blanchoin, L. & Mullins, R. D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 (2000).
Small, J. V. et al. Unravelling the structure of the lamellipodium. J. Microsc. 231, 479–485 (2008).
Aguilera, M. O., Beron, W. & Colombo, M. I. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy 8, 1590–1603 (2012).
Aplin, A., Jasionowski, T., Tuttle, D. L., Lenk, S. E. & Dunn, W. A. Cytoskeletabell elements are required for the formation and maturation of autophagic vacuoles. J. Cell Physiol. 152, 458–466 (1992).
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Barak, L. S., Yocum, R. R. & Webb, W. W. In vivo staining of cytoskeletal actin by autointernalization of nontoxic concentrations of nitrobenzoxadiazole-phallacidin. J. Cell Biol. 89, 368–372 (1981).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Riedl, J. et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008).
Derubeis, A. R., Young, M. F., Jia, L., Robey, P. G. & Fisher, L. W. Double FYVE-containing protein 1 (DFCP1): isolation, cloning and characterization of a novel FYVE finger protein from a human bone marrow cDNA library. Gene 255, 195–203 (2000).
Cheung, P. C., Trinkle-Mulcahy, L., Cohen, P. & Lucocq, J. M. Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. Biochem. J. 355, 113–121 (2001).
Ridley, S. H. et al. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 114, 3991–4000 (2001).
Gustafsson, M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Klionsky, D. J., Elazar, Z., Seglen, P. O. & Rubinsztein, D. C. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4, 849–950 (2008).
Hawkins, M., Pope, B., Maciver, S. K. & Weeds, A. G. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry 32, 9985–9993 (1993).
Hayden, S. M., Miller, P. S., Brauweiler, A. & Bamburg, J. R. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 32, 9994–10004 (1993).
Yonezawa, N., Nishida, E. & Sakai, H. pH control of actin polymerization by cofilin. J. Biol. Chem. 260, 14410–14412 (1985).
Moon, A. & Drubin, D. G. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol. Biol. Cell 6, 1423–1431 (1995).
Theriot, J. A. Accelerating on a treadmill: ADF/cofilin promotes rapid actin filament turnover in the dynamic cytoskeleton. J. Cell Biol. 136, 1165–1168 (1997).
Svitkina, T. M. & Borisy, G. G. Arp2/3 complex and actin depolymerizing factor cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026 (1999).
Yamaguchi, H. et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 168, 441–452 (2005).
Bailly, M. et al. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr. Biol. 11, 620–625 (2001).
Nolen, B. J. et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009).
Goley, E. D. & Welch, M. D. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 (2006).
Higgs, H. N. & Pollard, T. D. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70, 649–676 (2001).
Mullins, R. D., Heuser, J. A. & Pollard, T. D. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl Acad. Sci. USA 95, 6181–6186 (1998).
Welch, M. D., DePace, A. H., Verma, S., Iwamatsu, A. & Mitchison, T. J. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 138, 375–384 (1997).
Chhabra, E. S. & Higgs, H. N. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell. Biol. 9, 1110–1121 (2007).
Cooper, J. A. & Schafer, D. A. Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 12, 97–103 (2000).
Carlier, M. F. & Pantaloni, D. Control of actin assembly dynamics in cell motility. J. Biol. Chem. 282, 23005–23009 (2007).
Cooper, J. A. & Sept, D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267, 183–206 (2008).
Heiss, S. G. & Cooper, J. A. Regulation of CapZ, an actin capping protein of chicken muscle, by anionic phospholipids. Biochemistry 30, 8753–8758 (1991).
Hug, C., Miller, T. M., Torres, M. A., Casella, J. F. & Cooper, J. A. Identification and characterization of an actin-binding site of CapZ. J. Cell Biol. 116, 923–931 (1992).
Chen, R. et al. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J. Cell Biol. 206, 173–182 (2014).
Bjorkoy, G. et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 (2005).
Schafer, D. A., Jennings, P. B. & Cooper, J. A. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 (1996).
Liang, X. H. et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 (1999).
Furuya, N., Yu, F., Byfield, M., Pattingre, S. & Levine, B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 1, 46–52 (2005).
Monastyrska, I. et al. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell 19, 1962–1975 (2008).
Kast, D. J., Zajac, A. L., Holzbaur, E. L., Ostap, E. M. & Dominguez, R. WHAMM directs the Arp2/3 complex to the ER for autophagosome biogenesis through an actin comet tail mechanism. Curr. Biol. 25, 1791–1797 (2015).
Proikas-Cezanne, T., Takacs, Z., Donnes, P. & Kohlbacher, O. WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 128, 207–217 (2015).
Baskaran, S., Ragusa, M. J. & Hurley, J. H. How Atg18 and the WIPIs sense phosphatidylinositol 3-phosphate. Autophagy 8, 1851–1852 (2012).
Pollard, T. D. & Cooper, J. A. Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009).
Cooper, J. A. & Schafer, D. A. Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 12, 97–103 (2000).
Cooper, J. A. & Sept, D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267, 183–206 (2008).
Plutner, H., Davidson, H. W., Saraste, J. & Balch, W. E. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 119, 1097–1116 (1992).
Hayashi-Nishino, M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 (2009).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Schafer, D. A., Jennings, P. B. & Cooper, J. A. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 (1996).
Acknowledgements
We are grateful to Nikon instruments (Shanghai) and the Tsinghua Cell Biology Core Facility for providing technical support, and to Y. Li and L. Huang for assistance with confocal microscopy, TEM, and image processing. GFP–DFCP1 was a gift from N. T. Ktistakis. This research was supported by National Science Foundation Grants 31125018, 31030043 and 31321003, 973 Program 2010CB833704, 2011CB910100, and Tsinghua University Grants 2010THZ0 and 2009THZ03071 to L.Y.
Author information
Authors and Affiliations
Contributions
L.Y., Z.C. and N.M. conceived the idea. L.Y. supervised the study with help from N.M., Y.C. and Z.C. N.M. and Y.C. designed and conducted most of the experiments and analysed the data. N.G. and Q.G. helped with the tomography study. S.W., M.C., M.Z., G.Y., M.M., Q.S., S.L., J.S., Y.S. and J.X. contributed to the experiments. L.Y. and Y.C. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Actin puncta are co-localized with LC3 puncta.
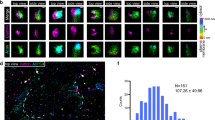
(a) GFP-LC3 expressing NRK cells were starved for 4 h and time-lapse images were acquired with a NIKON A1 confocal microscope. The number of autophagosomes at the indicated time points was quantified. (n = 3 independent experiments; 50 cells per time point were assessed per independent experiment.) Data represent mean ± s.d. (b) NRK cells were starved for 4 h then stained with phalloidin (to detect polymerized actin) and antibody to LC3. Regions of LC3 puncta that colocalize with phalloidin are outlined with white dashed lines and are magnified to the right. Scale bars in full panels and zoomed panels correspond to 5 μm and 1 μm, respectively.
Supplementary Figure 2 CapZ regulates autophagy.
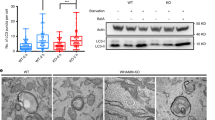
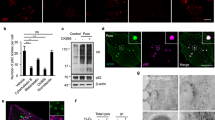
(a) RNAi knockdown efficiency for Capzb. Cells were transfected with non-specific (NS) RNAi or two different RNAis against the Capzb gene. 60 h after transfection, the Capzb mRNA level was measured by qPCR. Data shown are from one experiment. The experiment was repeated 3 times. (b) NRK cells were transfected with nonspecific- (NS) or Capzb-RNAi. Cells were starved for 0 or 2 h and analyzed by western blot with an antibody to actin or LC3. Uncropped images of blots are shown in Supplementary Fig. 9. (c) NRK cells were transfected with nonspecific- (NS) or Capzb-RNAi-2. Cells were starved for 4 h and stained with antibody to LC3. Scale bar, 5 μm. (d) Cells from (c) were assessed for abnormal tubular LC3 puncta in a blinded fashion and quantified. (n = 3 independent experiments; 50 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗P < 0.05 (two-tailed t-test). (e) GFP-LC3-expressing NRK cells were transfected with nonspecific- (NS) or Capzb-RNAi. Cells were starved for 4 h and stained with antibody to GFP. Cells were quantified for total LC3 puncta. (n = 3 independent experiments; 50 cells were assessed per independent experiment). Data represents mean ± s.d. NS, no significant (two-tailed t-test). (f) Raw cells were transfected with nonspecific- (NS) or Capzb-RNAi. Cells were starved for 4 h and stained with antibody to LC3. Scale bar, 5 μm. (g) Cells from (f) were assessed for abnormal tubular LC3 puncta in a blinded fashion and quantified. (n = 3 independent experiments; 100 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗∗∗P < 0.001 (two-tailed t-test). (h) Stable Capzb knockdown NRK cells were transfected with CapZβ-Myc. Cells were starved for 0 or 4 h and stained with antibody to LC3. Scale bar, 5 μm. (i) The expression level of CapZβ-Myc was verified by western blot. Uncropped images of blots are shown in Supplementary Fig. 9. (j) Cells from (h) were assessed for abnormal tubular LC3 puncta in a blinded fashion and quantified. (n = 3 independent experiments; 100 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗P < 0.05 (two-tailed t-test).
Supplementary Figure 3 CapZ regulates autophagy.
(a) GFP-DFCP1-expressing NRK cells were transfected with nonspecific- (NS) or Capzb-RNAi. Cells were starved for 4 h and stained with antibodies to GFP. Regions outlined with white dashed lines are magnified. Scale bars in full panels and zoomed panels correspond to 5 μm and 2 μm, respectively. (b) GFP-DFCP1-expressing NRK cells were transfected with nonspecific- (NS) or Capzb-RNAi. Cells were stained with antibodies to GFP and LC3. Scale bar, 5 μm.
Supplementary Figure 4 Formation of abnormal LC3 puncta in CK666-treated cells.
(a) NRK cells were starved for 2 h, then 100 μM CK666 was added to the starvation medium for 1 h. Cells were stained with antibody to LC3. Scale bar, 5 μm. (b) Cells from (a) were assessed for abnormal tubular LC3 puncta in a blinded fashion and quantified. (n = 3 independent experiments; 100 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗P < 0.05 (two-tailed t-test. (c) GFP-DFCP1-expressing NRK cells were starved for 2 h, then 100 μM CK666 was added to the starvation medium for 1 h. Cells were stained with antibodies to GFP and LC3. Scale bar, 5 μm. (d) Cells from (c) were assessed for DFCP1/LC3 double-positive puncta in a blinded fashion and quantified. (n = 3 independent experiments; 50 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗P < 0.05 (two-tailed t-test). (e) NRK cells were starved for 2 h, then 100 μM CK666 was added to the starvation medium for 1 h. Cells were observed by TEM. Scale bar, 1 μm. (f) Cells from (e) were assessed for abnormal autophagosomes in a blinded fashion and quantified. (n = 3 independent experiments; 60 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗∗P < 0.01 (two-tailed t-test).
Supplementary Figure 5 CapZ regulates actin puncta formation.
(a) NRK cells were transfected with nonspecific- (NS) or Capzb-RNAi. Cells were starved for 0 or 4 h and stained with phalloidin and antibody to LC3. Scale bar, 5 μm. (b) Cells from (a) were quantified for phalloidin-positive LC3 puncta. (n = 3 independent experiments; 50 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗∗P < 0.01 (two-tailed t-test). (c) Stable Capzb knockdown NRK cells were transfected with CapZβ-Myc, then starved for 0 or 4 h and stained with antibody to LC3 and phalloidin. Regions outlined with white dashed lines are magnified. Scale bars in full panels and zoomed panels correspond to 5 μm and 1 μm, respectively. (d) Cells from (c) were quantified for phalloidin-positive LC3 puncta. (n = 3 independent experiments; 100 cells were assessed per independent experiment). Data represent mean ± s.d. ∗∗∗P < 0.001 (two-tailed t-test).
Supplementary Figure 6
(a) CapZ mutant has impaired binding activity to PI3P. Co-sedimentation assay with PI3P micelles and recombinant WT CapZαCapZβ and CapZα(K256A, R260A)CapZβ(R225A). Proteins in supernatant (S) and precipitate (P) were visualized by SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. The experiment was repeated 3 times. Uncropped images of gels are shown in Supplementary Fig. 9. (b) GFP-DFCP1-expressing NRK cells were transfected with wild type or mutant CapZα(K256A, R260A)-Myc and CapZβ(R225A)-Myc. Cells were then starved for 2 h and stained with antibodies to GFP and Myc. Regions outlined with white dashed lines are magnified. Scale bars in full panels and zoomed panels correspond to 5 μm and 1 μm, respectively. (c) Cells from (b) were quantified for CapZ-positive DFCP1 puncta. (n = 3 independent experiments; 50 cells were assessed per independent experiment.) Data represent mean ± s.d. ∗∗P < 0.01 (two-tailed t-test).
Supplementary Figure 7 Co-sedimentation assay of CapZαCapZβ/Cofilin with liposomes containing 20% PI3P or PI(4,5)P2.
Proteins in supernatant (S) and precipitate (P) were visualized by SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. The experiment was repeated 3 times. Uncropped images of gels are shown in Supplementary Fig. 9.
Supplementary Figure 8
(a) Uncapping efficiency of PI3P is dose-dependent. Uncapping assays were performed with 2 μMα-actin and 10 nM CapZ. The indicated concentrations of PI3P were added to the reactions 20 min after the start point. Uncapping efficiency was measured by the increase in the rate of actin polymerization. (b) Comparison of the effects of PI(4,5)P2 and PI3P on uncapping of actin filaments. Uncapping assays were performed with 2 μMα-actin and 10 nM CapZ. 90 μM PI3P (cyan curve) or PI(4,5)P2 (blue curve) was added to the reactions. Both PI3P and PI(4,5)P2 induced a large increase in the rate of actin polymerization from capped actin filaments.
Supplementary Figure 9 Scans of original Western Blot analyses and SDS-PAGE followed by Coomassie brilliant blue analysis.
Cropped regions are indicated by the red boxes. Molecular weights are reported.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1387 kb)
The kinetic of serum/glutamine starvation induced autophagy.
GFP-LC3-expressing NRK cells were starved for 4 h in serum/glutamine starvation medium. Cells were observed by spinning disc microscope. Time interval, 10 min. (AVI 5677 kb)
Rights and permissions
About this article
Cite this article
Mi, N., Chen, Y., Wang, S. et al. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat Cell Biol 17, 1112–1123 (2015). https://doi.org/10.1038/ncb3215
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3215
This article is cited by
-
The temporal association of CapZ with early endosomes regulates endosomal trafficking and viral entry into host cells
BMC Biology (2024)
-
ARP2/3 complex associates with peroxisomes to participate in pexophagy in plants
Nature Plants (2023)
-
GRAF1 integrates PINK1-Parkin signaling and actin dynamics to mediate cardiac mitochondrial homeostasis
Nature Communications (2023)
-
Myosin 1D and the branched actin network control the condensation of p62 bodies
Cell Research (2022)
-
SPATA33 affects the formation of cell adhesion complex by interacting with CTNNA3 in TM4 cells
Cell and Tissue Research (2022)