Abstract
YAP/TAZ are nuclear effectors of the Hippo pathway regulating organ growth and tumorigenesis. Yet, their function as transcriptional regulators remains underinvestigated. By ChIP-seq analyses in breast cancer cells, we discovered that the YAP/TAZ transcriptional response is pervasively mediated by a dual element: TEAD factors, through which YAP/TAZ bind to DNA, co-occupying chromatin with activator protein-1 (AP-1, dimer of JUN and FOS proteins) at composite cis-regulatory elements harbouring both TEAD and AP-1 motifs. YAP/TAZ/TEAD and AP-1 form a complex that synergistically activates target genes directly involved in the control of S-phase entry and mitosis. This control occurs almost exclusively from distal enhancers that contact target promoters through chromatin looping. YAP/TAZ-induced oncogenic growth is strongly enhanced by gain of AP-1 and severely blunted by its loss. Conversely, AP-1-promoted skin tumorigenesis is prevented in YAP/TAZ conditional knockout mice. This work highlights a new layer of signalling integration, feeding on YAP/TAZ function at the chromatin level.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sudol, M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 9, 2145–2152 (1994).
Kanai, F. et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19, 6778–6791 (2000).
Piccolo, S., Dupont, S. & Cordenonsi, M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94, 1287–1312 (2014).
Ramos, A. & Camargo, F. D. The Hippo signaling pathway and stem cell biology. Trends Cell Biol. 22, 339–346 (2012).
Harvey, K. F., Zhang, X. & Thomas, D. M. The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013).
Halder, G. & Johnson, R. L. Hippo signaling: growth control and beyond. Development 138, 9–22 (2011).
Aragona, M. et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013).
Strano, S. et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA damage. Mol. Cell 18, 447–459 (2005).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Rosenbluh, J. et al. β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151, 1457–1473 (2012).
Yagi, R., Chen, L. F., Shigesada, K., Murakami, Y. & Ito, Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18, 2551–2562 (1999).
Calo, E. & Wysocka, J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013).
Rhie, S. et al. Nucleosome positioning and histone modifications define relationships between regulatory elements and nearby gene expression in breast epithelial cells. BMC Genomics 15, 331 (2014).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Cordenonsi, M. et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011).
Bhat, K. P. et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 25, 2594–2609 (2011).
Lamar, J. M. et al. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl Acad. Sci. USA 109, E2441–E2450 (2012).
Skibinski, A. et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 6, 1059–1072 (2014).
Eferl, R. & Wagner, E. F. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868 (2003).
E. P. Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Koos, B. et al. Analysis of protein interactions in situ by proximity ligation assays. Curr. Top. Microbiol. Immunol. 377, 111–126 (2014).
Shao, D. D. et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184 (2014).
Brown, P. H., Alani, R., Preis, L. H., Szabo, E. & Birrer, M. J. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene 8, 877–886 (1993).
Santner, S. J. et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 65, 101–110 (2001).
Bakiri, L., Matsuo, K., Wisniewska, M., Wagner, E. F. & Yaniv, M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell Biol. 22, 4952–4964 (2002).
Balmain, A. & Yuspa, S. H. Milestones in skin carcinogenesis: the biology of multistage carcinogenesis. J. Invest. Dermatol. 134, E2–E7 (2014).
Bailleul, B. et al. Skin hyperkeratosis and papilloma formation in transgenic mice expressing a ras oncogene from a suprabasal keratin promoter. Cell 62, 697–708 (1990).
Young, M. R. et al. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc. Natl Acad. Sci. USA 96, 9827–9832 (1999).
Briso, E. M. et al. Inflammation-mediated skin tumorigenesis induced by epidermal c-Fos. Genes Dev. 27, 1959–1973 (2013).
Zenz, R. et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev. Cell 4, 879–889 (2003).
Chen, Y. & Lai, M. Z. c-Jun NH2-terminal kinase activation leads to a FADD-dependent but Fas ligand-independent cell death in Jurkat T cells. J. Biol. Chem. 276, 8350–8357 (2001).
Azzolin, L. et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170 (2014).
Kapoor, A. et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158, 185–197 (2014).
Tam, W. L. et al. Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 24, 347–364 (2013).
Bakiri, L. et al. Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFbeta expression. Cell Death Differ. 22, 336–350 (2015).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Li, M. et al. Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature 407, 633–636 (2000).
Dupont, S. et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 (2009).
Martello, G. et al. A microRNA targeting dicer for metastasis control. Cell 141, 1195–1207 (2010).
Azzolin, L. et al. Role of TAZ as mediator of Wnt signaling. Cell 151, 1443–1456 (2012).
Schmidt, D. et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779 (2011).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Kharchenko, P. V., Tolstorukov, M. Y. & Park, P. J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26, 1351–1359 (2008).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Enzo, E. et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 34, 1349–1370 (2015).
Irizarry, R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003).
Dai, M. et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33, e175 (2005).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001).
Huang, F., He, J., Zhang, Y. & Guo, Y. Synthesis of biotin-AMP conjugate for 5’ biotin labeling of RNA through one-step in vitro transcription. Nat. Protoc. 3, 1848–1861 (2008).
Debnath, J., Muthuswamy, S. K. & Brugge, J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003).
Bodega, B. et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 7, 41 (2009).
Zhang, H. et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 284, 13355–13362 (2009).
Zhang, N. et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38 (2010).
Morsut, L. et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development 137, 2571–2578 (2010).
Adorno, M. et al. A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 (2009).
Acknowledgements
We thank O. Wessely, S. Dupont and G. Martello for comments; M. Morgante and E. Aleo for deep-sequencing (IGA, Udine); V. Guzzardo for histology; D. J. Pan (Johns Hopkins University, Maryland, USA) for gifts of Yap fl/fl mice; D. Metzger and P. Chambon (University of Strasbourg, France) for K14–Cre–ERT2 transgenics; L. Naldini (San Raffaele Scientific Institute, Italy) for lentiviral plasmids and L. Bakiri (Spanish National Cancer Research Centre (CNIO), Spain) for AP-1 expression constructs. We are grateful to C. Frasson for flow cytometry analyses. We acknowledge a donation in memoriam of L. Simonutti. F.Z. and L.A. are supported by a fellowship from the Italian Association for Cancer Research (AIRC). M.F. is supported by an assistant professorship from FIRB Accordi di Programma 2011 RBAP11T3WB. This work is supported by AIRC-MFAG to M.C.; from AIRC Special Program Molecular Clinical Oncology ‘5 per mille’ to S.P. and S.B.; Epigenetics Flagship project CNR-MIUR grants to S.B. and S.P.; AIRC-IG Grant to S.P. Work in the laboratory of S.P. is also supported by ERC-2014-ADG.
Author information
Authors and Affiliations
Contributions
F.Z. and G.B. performed experiments, M.F., M.C. and S.B. performed bioinformatic analysis, E.Q. helped with molecular biology, L.A. and A.R. performed in vivo experiments, B.B. helped with ChIP and 3C protocols, M.C. and S.P. planned, discussed and organized the work and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
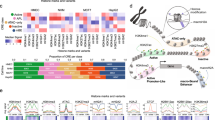
Supplementary Figure 1 Genome-wide identification of YAP/TAZ/TEAD binding sites.
(a) ChIP-qPCR on the promoters of established YAP/TAZ direct targets showing the specificity of YAP and TAZ antibodies. CTGF and CYR61 promoter sequences (but not a negative control locus) were enriched in YAP- and TAZ-immunoprecipitated chromatin, but not in negative control IP (IgG) or in chromatin obtained from YAP/TAZ-depleted cells. Relative DNA binding was calculated as fraction of input and normalized to IgG. Data from 2 biological replicates (individual data points and their mean) from one representative experiment are shown. (b,c) YAP/TAZ binding profiles and called peaks at positive control loci (b) and other known YAP/TAZ-regulated genes (c). Asterisks indicate peaks validated in (d). (d) Validation by ChIP-qPCR of YAP/TAZ binding sites identified through ChIP-seq. 2 biological replicates from one representative experiment are shown. (e) Percentage of YAP/TAZ peaks containing at least one known TEAD-binding motif. (f) Position of TEAD motif relative to the top of YAP/TAZ peaks. The curve shows the density of TEAD motifs at each position in a 500 bp window surrounding the summit of the corresponding YAP/TAZ peaks. (g) Specificity of TEAD4 antibody was assessed by ChIP-qPCR in control (siCO) and TEAD4-depleted MDA-MB-231 cells (siT4). 2 biological replicates (individual data points and their mean) from one representative experiment are shown. (h) ChIP-seq profiles, showing co-occupancy of CTGF and ANKRD1 promoters by YAP/TAZ and TEAD4. (i) Genomic distribution of TEAD4 or TEAD1 ChIP-seq peaks relative to the closest TSS in the indicated cell types. ERMS = embryonal rhabdomyosarcoma. (j) ChIP-qPCR comparing the levels of H3K4me3 and H3K4me1 (normalized to total H3 levels) in a group of YAP/TAZ/TEAD-bound promoters and enhancers. Data points are the mean of two biological replicates from one representative experiment. ‘Promoters’ are YAP/TAZ/TEAD peaks close to the TSS of ANKRD1, AMOTL2, AXL, TK1, AJUBA and WTIP; enhancers are YAP/TAZ/TEAD peaks 40841 (connected to MYC—see text and Supplementary Table 1), 40896 (MYC), 24736 (GINS1), 31079 (CCNA2), 16872, 16908, 16914 (TOP2A), 7673 (CDCA5), 7773 (FOSL1), 16878 (CDC6). (k) Percentage of YAP/TAZ/TEAD peaks localized in nucleosome-depleted regions, defined by FAIRE. (l) Schematic representation of the procedure used to identify candidate YAP/TAZ/TEAD direct target genes. See Methods for reproducibility of experiments.
Supplementary Figure 2 YAP/TAZ/TEAD cell proliferation program.
(a) mRNA levels (measured by qRT-PCR and normalized to GAPDH) for TEAD1-4 in MDA-MB-231 cells transfected with siTEAD A or siTEAD B. Data from 2 biological replicates (individual data points and their mean) from one representative experiment are shown. The effectiveness of TEAD depletion was also evaluated by qRT-PCR for the expression of the YAP/TAZ/TEAD targets CTGF and ANKRD1. (b) Western blot for the indicated proteins in MDA-MB-231 cells transfected with control siRNA (siCO), YAP/TAZ siRNAs (siYT) or TEAD siRNAs (siTEAD). GAPDH serves as loading control. (c) ChIP-qPCR verifying YAP/TAZ binding to the enhancers associated with a subset of YAP/TAZ target genes presented in Fig. 2b. Relative DNA binding was calculated as fraction of input and normalized to IgG; data from 2 biological replicates from one representative experiment are shown. See Supplementary Fig. 1d for negative control locus. (d) Validation of the DNA looping interaction between the MYC promoter and a downstream YAP/TAZ/TEAD-occupied enhancer, using 3C assay. Data are presented as in Fig. 2c. Data points are mean + s.e.m. from n = 3 biological replicates. (e) Validation of the DNA looping interaction between the TOP2A promoter and a downstream YAP/TAZ/TEAD-occupied enhancer, using 3C assay. Data are presented as in Fig. 2c. Data points are mean + s.e.m. from n = 3 biological replicates. See Methods for reproducibility of experiments.
Supplementary Figure 3 Control of cell proliferation by YAP/TAZ, TEAD and their target MYC.
(a) Growth of MDA-MB-231 cells transfected with control siRNA (siCO), YAP siRNA (siYAP), TAZ siRNA (siTAZ) or a combination of YAP and TAZ siRNAs (siYT). Data are mean + SD of n = 8 biological replicates. (b) Growth curve of MDA-MB-231 cells transfected with control siRNA (siCO) or TEAD siRNAs (siTEAD). Data are mean + SD of n = 8 biological replicates. (c) Percentage of MDA-MB-231 cells in G1, S and G2/M phases of cell cycle, as determined by flow-cytometric analysis. Cells were transfected with control (siCO) or TEAD siRNAs (siTEAD) 48 h before fixation. Data are mean + SD of n = 3 biological replicates. (d) Sustained expression of YAP, but not of TEAD-binding deficient YAPS94A, rescues cell proliferation in YAP/TAZ-depleted cells. Empty-vector-, wild-type YAP- (wt) or YAPS94A-transduced MDA-MB-231 cells were transfected with control (siCO) or YAP/TAZ (siYT) siRNAs, as indicated. Proliferation was evaluated as in (a). Data are mean + SD of n = 8 biological replicates. (e) Western blot showing Myc depletion in cells transfected with two MYC siRNAs. GAPDH serves as loading control. (f) Growth curve of MDA-MB-231 cells transfected with control siRNA (siCO) or MYC siRNAs (siMYC). Data are mean + SD of n = 8 biological replicates. (g) Percentage of MDA-MB-231 cells in G1, S and G2/M phases of the cell cycle, as determined by flow-cytometric analysis. Cells were transfected with control (siCO) or MYC siRNAs (siMYC) 48h before fixation. Data are mean + SD of n = 3 biological replicates. (h) MDA-MB-231 cells were transduced with lentiviral vectors encoding rtTA and doxycycline-inducible EGFP (MDA TetON EGFP) and transfected with control or YAP/TAZ siRNAs. Where indicated, EGFP expression was induced with 0.1 μg ml−1 doxycycline at the time of transfection. Cell growth was evaluated as in (a). Data are mean + SD of n = 8 biological replicates. (i) Correlation between the expression of validated YAP/TAZ/TEAD (YTT) direct target gene signature and TAZ (WWTR1) mRNA or established YAP/TAZ signatures in breast cancer samples. Pearson correlation coefficients (r) and P-values are shown (one-tailed t test, P < 0.0001 for all correlations). See Methods for reproducibility of experiments.
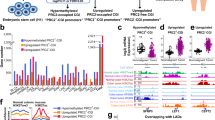
Supplementary Figure 4 Chromatin co-occupancy of YAP, TAZ, TEAD and AP-1 at the genome-wide level.
(a) Western blot showing the specificity of antibodies recognizing AP-1 proteins used in this study. GAPDH serves as loading control. (b) Left: ChIP-qPCR on the promoter of the established AP-1 direct target FOSL1 showing the specificity of JUN antibody. FOSL1 promoter sequence was enriched in JUN-immunoprecipitated chromatin, but not in negative control IP (IgG) or in chromatin obtained from JUN-depleted cells. Relative DNA binding was calculated as fraction of input and normalized to IgG; data are presented as mean + SD of n = 3 biological replicates. Right: Western blot showing that JUN is specifically immunoprecipitated by JUN antibody from crosslinked chromatin. (c) JUN binding profiles at FOSL1 locus and known YAP/TAZ-regulated genes. (d) ChIP-qPCR validating JUN binding to the indicated genomic regions. Data from 2 biological replicates (individual data points and their mean) from one representative experiment are shown. (e) Left: ChIP-qPCR on the promoter of the established AP-1 direct target FOSL1 showing the specificity of FOSL1 (R-20) antibody. Right: ChIP-qPCR showing FOSL1 binding to the same genomic regions occupied by JUN, as in (c). ‘siFL1’ indicates cells transfected with FOSL1 siRNAs (serving as negative control). Data from 2 biological replicates (individual data points and their mean) from one representative experiment are shown. (f) Percentage of TEAD4 peaks overlapping with at least a ChIP-seq peak for a member of JUN and FOS families in MDA-MB-231 cells and in other cancer cell lines. (g) Heatmap representing TEAD4, JUND and FOSL2 ChIP-seq reads in HepG2 cells. TEAD4 binding sites are ranked from the strongest to weakest signal; a window of ±1 kb centered on the summit of TEAD4 peaks is shown. See Methods for reproducibility of experiments.
Supplementary Figure 5 Biochemical and functional interactions between YAP/TAZ/TEAD and AP-1 proteins.
(a,b) In situ PLA detection of endogenous YAP/AP-1 and TEAD1/AP-1 interactions in MDA-MB-231 cells (a), HCT116 cells (b, left) and A549 cells (b, right). See Supplementary Table 6 for details about antibodies. Magnification is the same for all pictures. (c) TEAD1 co-precipitates with FOSL1 at endogenous protein levels in HCT116 cells. Two FOSL1 antibodies (N-17 and R-20) were used for IP. JUN is a positive control for co-IP. All samples were run on the same gel. (d) TEAD1, but not YAP, co-precipitates with FOSL1 and JUND immunocomplexes purified from MDA-MB-231 cells. Two independent immunoprecipitations were performed with FOSL1 (N-17) and JUND antibodies, or with control IgGs. Lanes 4 and 5 are also presented in Fig. 3h; all samples were run on the same gel. (e) JUN and JUND cannot be detected in YAP immunocomplexes purified from MDA-MB-231 cells. TEAD1 is a positive control for co-IP. All samples were run on the same gel. (f) JUN and JUND cannot be detected in FLAG-YAP or FLAG-TAZ immunocomplexes purified from MDA-MB-231 cells. TEAD1 is a positive control for co-IP. (g) Linear correlation between the signal of YAP/TAZ peaks and TEAD4 or JUN peaks in the common binding sites. r2 is the coefficient of determination of the correlation. (h) AP-1 luciferase reporter (AP-1 luc) is activated by treatment with TPA in HEK293 cells. Data are mean + SD of n = 4 biological replicates from 2 independent experiments. (i) HEK293 cells were transfected with increasing doses of YAP-expressing vector, and with an AP-1 (pAP1-luc, left) or TEAD (8xGTIIC-LUX, right) luciferase reporter. Data are normalized to control sample (‘-’) and are presented as mean + SD of n = 4 biological replicates from 2 independent experiments. (j) AP-1 or TEAD fail to bind to CTGF promoter after mutation of their cognate binding sites. Panels are Western blot analyses of the indicated endogenous proteins from MDA-MB-231 nuclear extracts, purified by DNA-pull down using biotinylated DNA probes designed on the sequence of CTGF promoter. See Supplementary Fig. 7 for uncropped Western blots, and Methods for reproducibility of experiments.
Supplementary Figure 6 Role of YAP/TAZ and AP-1 in oncogenic growth.
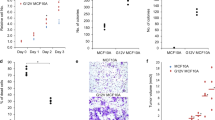
(a) MDA-MB-231 cells were transduced with rtTA and doxycycline inducible JUN-DN (MDA TetON JUN-DN); clones were established to ensure high and uniform expression of JUN-DN upon doxycycline treatment. Similarly, clones of EGFP-expressing cells were generated as control (MDA TetON EGFP). Levels of the indicated transcripts were evaluated by qRT-PCR in cells untreated or exposed to doxycycline for 48h. mRNA levels are normalized to GAPDH. Data from 2 biological replicates (individual data points and their mean) from one representative experiment are shown. Downregulation of FOSL1 transcript is a positive control for inhibition of AP-1 activity. ANKRD1 and CTGF expression diminishes in doxycycline-treated MDA TetON JUN-DN cells, coherently with results of luciferase reporters in Fig. 4i. (b) Growth rate of MDA TetON JUN-DN cells decreases upon JUN-DN induction with doxycycline (left panel); as control, EGFP induction has no effect (right panel). Data are mean + SD of n = 8 biological replicates. (c) Control and TAZS89A-overexpressing MII cells were transfected with the indicated siRNAs and tested for mammosphere formation. Data are presented as mean + SD of n = 6 biological replicates from a representative experiment. (d) Genomic DNA was extracted from the skin of control mice (n = 9, left) and conditional knockout mice (n = 9, right) to verify tamoxifen-induced recombination of Yap and Taz loci. Panels are PCR bands for the indicated alleles. (e) Magnifications of representative H&E-stained sections of skin tumors shown in Fig. 6b. Left: detail of a SCC area in a tumor of treated control mice. Right: DMBA/TPA-treated skin from YAP/TAZ conditional knockout mice has a normal histological appearance. See Methods for reproducibility of experiments.
Supplementary Figure 7 Uncropped Western blots.
Uncropped images of immunoblots displayed in the main and Supplementary figures. Dashed boxes indicate areas that were cropped.
Supplementary information
Supplementary Information
Supplementary Information (PDF 6388 kb)
Supplementary Table 1
Supplementary Information (XLSX 210 kb)
Supplementary Table 2
Supplementary Information (XLSX 616 kb)
Supplementary Table 3
Supplementary Information (XLSX 37 kb)
Supplementary Table 4
Supplementary Information (XLSX 14 kb)
Supplementary Table 5
Supplementary Information (XLSX 33 kb)
Supplementary Table 6
Supplementary Information (XLSX 46 kb)
Supplementary Table 7
Supplementary Information (XLSX 32 kb)
Supplementary Table 8
Supplementary Information (XLSX 45 kb)
Supplementary Table 9
Supplementary Information (XLSX 58 kb)
Rights and permissions
About this article
Cite this article
Zanconato, F., Forcato, M., Battilana, G. et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17, 1218–1227 (2015). https://doi.org/10.1038/ncb3216
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3216
This article is cited by
-
Matrix stiffness affects tumor-associated macrophage functional polarization and its potential in tumor therapy
Journal of Translational Medicine (2024)
-
Alternative Wnt-signaling axis leads to a break of oncogene-induced senescence
Cell Death & Disease (2024)
-
Inhibition of the YAP-MMB interaction and targeting NEK2 as potential therapeutic strategies for YAP-driven cancers
Oncogene (2024)
-
VAV2 orchestrates the interplay between regenerative proliferation and ribogenesis in both keratinocytes and oral squamous cell carcinoma
Scientific Reports (2024)
-
Tankyrase inhibition interferes with junction remodeling, induces leakiness, and disturbs YAP1/TAZ signaling in the endothelium
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)