Abstract
Throughout development, tissues undergo complex morphological changes, resulting from cellular mechanics that evolve over time and in three-dimensional space. During Drosophila germ-band extension (GBE), cell intercalation is the key mechanism for tissue extension1, and the associated apical junction remodelling is driven by polarized myosin-II-dependent contraction2,3,4. However, the contribution of the basolateral cellular mechanics to GBE remains poorly understood. Here, we characterize how cells coordinate their shape from the apical to the basal side during rosette formation, a hallmark of cell intercalation. Basolateral rosette formation is driven by cells mostly located at the dorsal/ventral part of the rosette (D/V cells). These cells exhibit actin-rich wedge-shaped basolateral protrusions and migrate towards each other. Surprisingly, the formation of basolateral rosettes precedes that of the apical rosettes. Basolateral rosette formation is independent of apical contractility, but requires Rac1-dependent protrusive motility. Furthermore, we identified Src42A as a regulator of basolateral rosette formation. Our data show that in addition to apical contraction, active cell migration driven by basolateral protrusions plays a pivotal role in rosette formation and contributes to GBE.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
05 March 2018
In the version of this Article originally published, the authors cited the wrong articles for reference numbers 18, 30 and 31; the correct ones are listed below. Furthermore, four additional references have been inserted at numbers 37, 38, 39 and 40 as in the list below, and the original references 37–40 have been renumbered. These corrections have been made in the online versions of the Article.
References
Irvine, K. D. & Wieschaus, E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827–841 (1994).
Bertet, C., Sulak, L. & Lecuit, T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667–671 (2004).
Zallen, J. A. & Wieschaus, E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343–355 (2004).
Blankenship, J. T., Backovic, S. T., Sanny, J. S., Weitz, O. & Zallen, J. A. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell 11, 459–470 (2006).
Collinet, C., Rauzi, M., Lenne, P.-F. & Lecuit, T. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nat. Cell Biol. 17, 1247–1258 (2015).
Yu, J. C. & Fernandez-Gonzalez, R. Local mechanical forces promote polarized junctional assembly and axis elongation in Drosophila. eLife 5, e10757 (2016).
Simoes Sde, M. et al. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell 19, 377–388 (2010).
Rauzi, M., Lenne, P.-F. & Lecuit, T. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 468, 1110–1114 (2010).
Tamada, M., Farrell, D. L. & Zallen, J. A. Abl regulates planar polarized junctional dynamics through β-catenin tyrosine phosphorylation. Dev. Cell 22, 309–319 (2012).
Levayer, R. & Lecuit, T. Oscillation and polarity of E-cadherin asymmetries control actomyosin flow patterns during morphogenesis. Dev. Cell 26, 162–175 (2013).
De Matos Simões, S., Mainieri, A. & Zallen, J. A. Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J. Cell Biol. 204, 575–589 (2014).
Britton, J. S., Lockwood, W. K., Li, L., Cohen, S. M. & Edgar, B. A. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239–249 (2002).
Pickering, K., Alves-Silva, J., Goberdhan, D. & Millard, T. H. Par3/Bazooka and phosphoinositides regulate actin protrusion formation during Drosophila dorsal closure and wound healing. Development 140, 800–809 (2013).
Williams-Masson, E. M., Heid, P. J., Lavin, C. A. & Hardin, J. The cellular mechanism of epithelial rearrangement during morphogenesis of the Caenorhabditis elegans dorsal hypodermis. Dev. Biol. 204, 263–276 (1998).
Walck-Shannon, E., Reiner, D. & Hardin, J. Polarized Rac-dependent protrusions drive epithelial intercalation in the embryonic epidermis of C. elegans. Development 1, 3549–3560 (2015).
Walck-Shannon, E. et al. CDC-42 orients cell migration during epithelial intercalation in the Caenorhabditis elegans epidermis. PLoS Genet. 12, e1006415 (2016).
Munro, E. M. & Odell, G. M. Polarized basolateral cell motility underlies invagination and convergent extension of the ascidian notochord. Development 129, 13–24 (2002).
Shih, J. & Keller, R. Cell motility driving mediolateral intercalation in explants of Xenopus laevis. Development 116, 901–914 (1992).
Krause, M. & Gautreau, A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15, 577–590 (2014).
Abreu-Blanco, M. T., Verboon, J. M. & Parkhurst, S. M. Coordination of Rho family GTPase activities to orchestrate cytoskeleton responses during cell wound repair. Curr. Biol. 24, 144–155 (2014).
Wu, Y. I. et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 (2009).
Wang, X., He, L., Wu, Y. I., Hahn, K. M. & Montell, D. J. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12, 591–597 (2010).
Martin, A. C., Kaschube, M. & Wieschaus, E. F. Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–499 (2009).
Fernandez-Gonzalez, R., Simoes Sde, M., Roper, J. C., Eaton, S. & Zallen, J. A. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell 17, 736–743 (2009).
Karess, R. E. et al. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell 65, 1177–1189 (1991).
Kiehart, D. P. Molecular genetic dissection of myosin heavy chain function. Cell 60, 347–350 (1990).
Chou, T. B. & Perrimon, N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131, 643–653 (1992).
Takahashi, M., Takahashi, F., Ui-Tei, K., Kojima, T. & Saigo, K. Requirements of genetic interactions between Src42A, armadillo and shotgun, a gene encoding E-cadherin, for normal development in Drosophila. Development 132, 2547–2559 (2005).
Brunet, T. et al. Evolutionary conservation of early mesoderm specification by mechanotransduction in Bilateria. Nat. Commun. 4, 2821 (2013).
Shih, J. & Keller, R. Patterns of cell motility in the organizer and dorsal mesoderm of Xenopus laevis. Development 116, 915–930 (1992).
Jessen, J. R. et al. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 4, 610–615 (2002).
Keller, R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science 298, 1950–1954 (2002).
Pare, A. C. et al. A positional toll receptor code directs convergent extension in Drosophila. Nature 515, 523–527 (2014).
Singh, J., Aaronson, S. A. & Mlodzik, M. Drosophila Abelson kinase mediates cell invasion and proliferation through two distinct MAPK pathways. Oncogene 29, 4033–4045 (2010).
Shindo, M. et al. Dual function of Src in the maintenance of adherens junctions during tracheal epithelial morphogenesis. Development 135, 1355–1364 (2008).
Förster, D. & Luschnig, S. Src42A-dependent polarized cell shape changes mediate epithelial tube elongation in Drosophila. Nat. Cell Biol. 14, 526–534 (2012).
Shindo, A. & Wallingford, J. B. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science 343, 649–652 (2014).
Williams, M., Yen, W., Lu, X. & Sutherland, A. Distinct apical and basolateral mechanisms drive planar cell polarity-dependent convergent extension of the mouse neural plate. Dev. Cell 29, 34–46 (2014).
Walck-Shannon, E. & Hardin, J. Cell intercalation from top to bottom. Nat. Rev. Mol. Cell Biol. 15, 34–48 (2014).
Shindo, A. Models of convergent extension during morphogenesis. WIREs Dev. Biol. 7, e293 (2018).
Foe, V. E., Field, C. M. & Odell, G. M. Microtubules and mitotic cycle phase modulate spatiotemporal distributions of F-actin and myosin II in Drosophila syncytial blastoderm embryos. Development 127, 1767–1787 (2000).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Mayer, M. et al. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature 467, 617–621 (2010).
Doncheva, N. T., Assenov, Y., Domingues, F. S. & Albrecht, M. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 7, 670–685 (2012).
Acknowledgements
We thank T. Lecuit (Institut de Biologie du Développement de Marseille, France), and the Bloomington Stock Center for fly stocks. We acknowledge S. Hayashi (RIKEN Center for Developmental Biology, Japan) for the generous gift of pSrc antibody. We thank the Microscopy Core of Mechanobiology Institute for imaging. We also thank M. Sheetz, S. Yonemura, R. Zaidel-Bar and MBI Science Communication Core (S. Wolf, and A. Wong) for helpful discussions and critical reading of the manuscript. We thank H. T. Ong for help in image quantifications. T.E.S. was supported by the National Research Foundation Singapore under its NRF Fellowship (NRF2012NRF-NRFF001-094). This work was supported by Mechanobiology Institute and National University of Singapore Startup Grants (to Y.T.), Temasek Life Sciences Laboratory (to Y.T.), and a Singapore Ministry of Education Tier 2 grant (MOE2015-T2-1-116 to Y.T.).
Author information
Authors and Affiliations
Contributions
Z.S. and Y.T. designed the experiments and wrote the manuscript. Z.S. performed the experiments. Z.S. and Y.T. analysed the data. M.S. and Y.H. contributed quantitative analyses. Z.S. and C.A. generated the plasmids. C.A. and T.E.S. contributed new reagents. Y.T. oversaw the project. All the authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
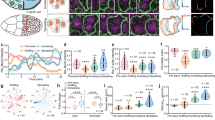
Supplementary Figure 1 Characteristics of rosette formation and GBE in wild-type and DN-Rac1T17N embryos.
(a) Forced-colored confocal time-lapse images of embryo expressing RFP::CAAX during rosette formation. Brighter colors denote high fluorescent intensity. D/V and A/P cells are indicated by filled and open dots, respectively. 0 min represents the time of rosette pattern formation. (a′) Quantification of RF/B of D/V cells in embryos expressing RFP::CAAX before and during rosette formation at the apical and a basal section (n = 10 D/V cells from 2 embryos). (b) Two-photon time-lapse images of embryos expressing Utrophin::GFP during rosette formation at the apical (0 μm) and the basal (−15 μm) section. Colored dots indicate cells in a rosette. Red arrows indicate the migration direction of intercalating D/V cells. 0sec represents the beginning of imaging. (c) Time progression of front-to-back ratio RF/B of pak3RBD::GFP intensity measured during the rosette formation at basal section (n = 6 rosettes, 2 embryos). (d) Tissue extension of wild-type (n = 19) and tissue-level expression of DN-RacTN17 embryos (n = 44). Average values are shown as thick horizontal lines. (d′) Schematic illustration of the tissue extension measurement. The length between the posterior end of germband and the posterior end of the embryo (Dgmb, black arrow) is divided by the length between the cephalic furrow and the end of embryo (Dcph, red arrow). (e,f) Forced-colored two-photon images of pak3RBD::GFP (e) and GFP::Rac1 (f) during fast GBE stage of ventrolateral epithelium in WT embryos. Data shown represent mean ± s.e.m. in a′, and c. Statistical tests were done by Student’s t-test.∗ and ∗∗∗ denote P < 0.05 and P < 0.001, respectively. NS: not significant. Scale bars, 10 μm.
Supplementary Figure 2 Quantifications of H-1152, Y-27632 injections and sqh, zip RNAi experiments and analysis of rosette topology transition and basal protrusions.
(a) Rosette formation with H1152 injection. (b) Fraction of rosettes with H1152 treated (56 rosettes, 3 embryos) and sqh RNAi (118 rosettes, 6 embryos) embryos. (c) Clustering coefficient of rosette progression in H1152 treatment (n = 5 rosettes from 3 embryos). (d,e) Rosette formation in zip RNAi (d) and sqh RNAi embryos (e). In a,d,e, filled and open dots indicated D/V and A/P cells, respectively. In d,e, the data are representative of 5 rosettes per condition. Red open circles denote the timing of rosette formation (0 min). (f) Fraction of D/V cells with protrusions in control, H1152 injected, and sqh RNAi embryos. (g,g′) Distribution of the orientation of basal protrusion in H1152 injected (n = 46), and sqh RNAi (n = 36) embryos. A, P, D, and V denote anterior, posterior, dorsal and ventral direction, respectively. (h) Speed of D/V cell at the basal section (−15 μm, n = 7,6,5 rosettes in wild-type, H1152 injected and sqh RNAi embryos, respectively). (i) Number of rosettes appearing in wild-type, Y-27632 injected, H1152 injected, zip RNAi, and sqh RNAi embryos. (j) Graphical illustration of the topological stages of 6-cell rosette. Three topologically distinct stages are pre-rosette, rosette, and resolved stages. Red lines indicate the connections between cells among the rosette. (k) Graphical representations of cell connections among the rosette (red lines). Centers of each cell are represented by numbered nodes. (l) Table of clustering coefficient and parameters measured by Cytoscape. (m,n) Examples of the modified ‘kissing-circle’ algorithm to detect protrusions. (m) A cell was considered as a protruding cell. There are 6 circles within the cell, and the diameter of each circle is less than 40% of the larger circle. (n) A cell was considered as a non-protruding cell. There are only 2 circles within the cell, and the diameter of smaller circle is 50% of the largest circle. Data shown represent mean ± s.e.m. in c, h and i. Statistical tests were done by Student’s t-test in h. ∗ and ∗∗∗ denote P < 0.05 and P < 0.001, respectively. Scale bars, 10 μm.
Supplementary Figure 3 Analyses of Src42A and Src64B knockdowns.
(a) Example of severe epithelium defects in strong Src42A RNAi embryos at stage 6 and stage 7. Red arrowheads indicate the gaps between epithelia cells. Yellow dots indicate the cells precociously delaminated from epithelium during cellularization. (b,c) Two-photon images of a rosette progression in Src42A dsRNA injected (b) or dominant-negative DN-Src42A induced embryos (c). (d,e) Forced-colored time-lapse images of pak3RBD::GFP (d) and GFP::Rac1 (e) during rosette formation in Src42A dsRNA injected embryos. In b–e, coloured dots label cells among a rosette, 0 min represents the time when apical rosette forms. (f) Fold change of Src42A and Src64B mRNA in Src42A RNAi, Src64B RNAi (left and middle bars, induced by GAL4-UAS), and Src42A dsRNA injected embryos relative to the control (UAS-Src42A, UAS-Src64B corresponding to each group, and Oregon-R as control for injections, n = 3 independent biological replicates for each group). Please refer to Supplementary Table 4 for details. (g) Number of apical-rosettes appearing in wild-type (n = 146 rosettes from 7 embryos) and Src42A RNAi (n = 167 rosettes from 7 embryos) embryos. (h) Fraction of different rosette phenotypes in wild-type (146 rosettes, from 7 embryos), Src42A dsRNA injected (57 rosettes from 6 embryos), Src64B dsRNA injected (29 rosettes from 4 embryos), and co-injection of Src42A and Src64B dsRNA embryos (35 rosettes from 6 co-injected embryos). The fraction of ‘basal-first’ and ‘apical-first’ rosettes in Src64B injected embryos was similar to that of wild-type. Moreover, the fraction in the embryos with double suppression of Src42A and Src64B did not exacerbate the basal defects compared with single suppression of Src42A. (i) Extension of germband in wild-type (n = 19) and Src64B RNAi (n = 25) embryos. The degree of GBE delay in Src64B RNAi was to a lesser extent compared to that of Src42A RNAi (Fig. 5k). These data indicated that Src64B did not play a major role in basal rosette formation and GBE. Data shown represent mean ± s.e.m. in f and g. Statistical tests were done by Student’s t-test in g and i. ∗ denotes P < 0.05. Scale bars, 10 μm.
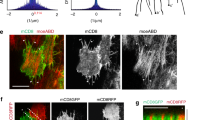
Supplementary Figure 4 pSrc and actin localization and the mesoderm morphology in wild-type and Src42A knockdown embryos.
(a,b) pSrc and phalloidin at the apical (a) and basolateral section (b) of a wild-type embryo. (a′, b′) High magnification of the boxed region in a and b. (b′) pSrc enriched at the basal rosette and co-localized with actin. (c-c′) Lateral view of pSrc and actin at GBE stage 7-8 ventral epithelium from wild-type, Oregon-R (c) and Src42A RNAi knockdown (c′) embryos. (d–d′) Cross-sectional view of mesoderm at stage 7-8 gastrulation embryos. The tissue integrity of invaginated mesoderm maintains in Src42A RNAi knockdown (d′) similar to the wild-type (d) embryo. (e) Distribution of pSrc and phalloidin of a basal defective rosette in DN-Src42A embryo. In a–a′, b–b′, c–c′, d–d′ images are representative of 4-5 embryos for each panel. In e, image is representative of more than 5 rosettes. Scale bars, 10 μm.
Supplementary Figure 5 Characteristics of basal intercalation during T1 transition, and analyses of rosette defects in eve, and Toll-2, 6, 8 knockdowns.
(a) Fractions of T1 transitions with ‘apical-first’ (white), ‘basal-first’ (light-grey), ‘simultaneous’ (dark-grey) phenotypes in wild-type embryos (n = 45 T1 transitions from 3 embryos). (b) Confocal time-lapse images of embryo expressing paGFP::Utr during T1 transition. Only small number of cells were photoactivated. D/V and A/P cells are indicated by filled and open dots, respectively. 0 min represents the time of the formation of basal new junction. White arrowheads indicate the direction of two intercalating D/V cells during T1 transition. White dotted lines represent the shape of cells with weaker fluorescence. (c) Quantifications of front-to-back ratio RF/B of D/V cells in embryos expressing pak3RBD::GFP (n = 10 D/V cells) and GFP::Rac1 (n = 16 D/V cells) intensity before and during T1 transition at the apical and a basal section. (d) Average number of rosettes counted at the apical section (0 μm) and a basal section (−15 μm) in wild-type (n = 4 embryos), eve RNAi (n = 9 embryos) and triple-knockdown of Toll-2,6,8 embryos (n = 6 embryos). Scale bar, 10 μm.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2703 kb)
Supplementary Table 1
Supplementary Information (XLSX 42 kb)
Supplementary Table 2
Supplementary Information (XLSX 47 kb)
Supplementary Table 3
Supplementary Information (XLSX 45 kb)
Supplementary Table 4
Supplementary Information (XLSX 155 kb)
41556_2017_BFncb3497_MOESM91_ESM.mov
Rosette formation at different apicobasal sections in wild-type embryo from two-photon live imaging of Utr-GFP. (MOV 261 kb)
Brightfield movie of GBE defect of an embryo expressing both zip RNAi and Src42A RNAi.
The embryo is representative from over 9 defective embryos. (MOV 2085 kb)
41556_2017_BFncb3497_MOESM97_ESM.mov
eve dsRNA injected embryo expressing Utr-GFP during GBE by two-photon imaging of apical and basal section. (MOV 5074 kb)
41556_2017_BFncb3497_MOESM98_ESM.mov
Toll-2,6,8 triple knockdown embryo expressing Utr-GFP during GBE by two-photon imaging of apical and basal section. (MOV 5105 kb)
Rights and permissions
About this article
Cite this article
Sun, Z., Amourda, C., Shagirov, M. et al. Basolateral protrusion and apical contraction cooperatively drive Drosophila germ-band extension. Nat Cell Biol 19, 375–383 (2017). https://doi.org/10.1038/ncb3497
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3497
This article is cited by
-
Basal actomyosin pulses expand epithelium coordinating cell flattening and tissue elongation
Nature Communications (2024)
-
Two Rac1 pools integrate the direction and coordination of collective cell migration
Nature Communications (2022)
-
Programmed and self-organized flow of information during morphogenesis
Nature Reviews Molecular Cell Biology (2021)
-
Tissue fluidity mediated by adherens junction dynamics promotes planar cell polarity-driven ommatidial rotation
Nature Communications (2021)
-
Looking deeper into tissue elongation
Nature Reviews Molecular Cell Biology (2020)