Abstract
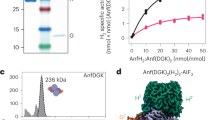
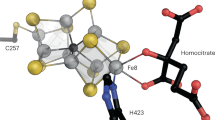
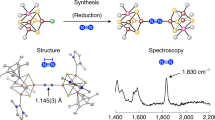
Nitrogenases catalyze the reduction of dinitrogen (N2) gas to ammonium at a complex heterometallic cofactor. This most commonly occurs at the FeMo cofactor (FeMoco), a [Mo–7Fe–9S–C] cluster whose exact reactivity and substrate-binding mode remain unknown. Alternative nitrogenases replace molybdenum with either vanadium or iron and differ in reactivity, most prominently in the ability of vanadium nitrogenase to reduce CO to hydrocarbons. Here we report the 1.35-Å structure of vanadium nitrogenase from Azotobacter vinelandii. The 240-kDa protein contains an additional α-helical subunit that is not present in molybdenum nitrogenase. The FeV cofactor (FeVco) is a [V–7Fe–8S–C] cluster with a homocitrate ligand to vanadium. Unexpectedly, it lacks one sulfide ion compared to FeMoco, which is replaced by a bridging ligand, likely a μ-1,3-carbonate. The anion fits into a pocket within the protein that is obstructed in molybdenum nitrogenase, and its different chemical character helps to rationalize the altered chemical properties of this unique N2- and CO-fixing enzyme.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Rees, D.C. Dinitrogen reduction by nitrogenase—If N2 isn't broken, it can't be fixed. Curr. Opin. Struct. Biol. 3, 921–928 (1993).
Rees, D.C. & Howard, J.B. Nitrogenase: standing at the crossroads. Curr. Opin. Chem. Biol. 4, 559–566 (2000).
Seefeldt, L.C., Hoffman, B.M. & Dean, D.R. Mechanism of Mo-dependent nitrogenase. Annu. Rev. Biochem. 78, 701–722 (2009).
Howard, J.B. & Rees, D.C. Structural basis of biological nitrogen fixation. Chem. Rev. 96, 2965–2982 (1996).
Eady, R.R. Structure–function relationships of alternative nitrogenases. Chem. Rev. 96, 3013–3030 (1996).
Einsle, O. Nitrogenase FeMo cofactor: an atomic structure in three simple steps. J. Biol. Inorg. Chem. 19, 737–745 (2014).
Bishop, P.E., Jarlenski, D.M.L. & Hetherington, D.R. Evidence for an alternative nitrogen fixation system in Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA 77, 7342–7346 (1980).
Bishop, P.E., Hawkins, M.E. & Eady, R.R. Nitrogen fixation in molybdenum-deficient continuous culture by a strain of Azotobacter vinelandii carrying a deletion of the structural genes for nitrogenase (nifHDK). Biochem. J. 238, 437–442 (1986).
Bishop, P.E. et al. Nitrogen fixation by Azotobacter vinelandii strains having deletions in structural genes for nitrogenase. Science 232, 92–94 (1986).
Robson, R.L. et al. The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme. Nature 322, 388–390 (1986).
Hales, B.J., Case, E.E., Morningstar, J.E., Dzeda, M.F. & Mauterer, L.A. Isolation of a new vanadium-containing nitrogenase from Azotobacter vinelandii. Biochemistry 25, 7251–7255 (1986).
Eady, R.R., Robson, R.L., Richardson, T.H., Miller, R.W. & Hawkins, M. The vanadium nitrogenase of Azotobacter chroococcum. Purification and properties of the VFe protein. Biochem. J. 244, 197–207 (1987).
Joerger, R.D. et al. Nucleotide sequences and mutational analysis of the structural genes for nitrogenase 2 of Azotobacter vinelandii. J. Bacteriol. 172, 3400–3408 (1990).
Eady, R.R. Current status of structure function relationships of vanadium nitrogenase. Coord. Chem. Rev. 237, 23–30 (2003).
Lee, C.C., Hu, Y. & Ribbe, M.W. Unique features of the nitrogenase VFe protein from Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA 106, 9209–9214 (2009).
Miller, R.W. & Eady, R.R. Molybdenum and vanadium nitrogenases of Azotobacter chroococcum. Low temperature favours N2 reduction by vanadium nitrogenase. Biochem. J. 256, 429–432 (1988).
Lee, C.C., Hu, Y. & Ribbe, M.W. Vanadium nitrogenase reduces CO. Science 329, 642 (2010).
Zhang, Q., Cheng, K., Kang, J., Deng, W. & Wang, Y. Fischer-Tropsch catalysts for the production of hydrocarbon fuels with high selectivity. ChemSusChem 7, 1251–1264 (2014).
Hu, Y., Lee, C.C. & Ribbe, M.W. Extending the carbon chain: hydrocarbon formation catalyzed by vanadium/molybdenum nitrogenases. Science 333, 753–755 (2011).
Hu, Y., Lee, C.C. & Ribbe, M.W. Vanadium nitrogenase: a two-hit wonder? Dalton Trans. 41, 1118–1127 (2012).
Hwang, J.C., Chen, C.H. & Burris, R.H. Inhibition of nitrogenase-catalyzed reductions. Biochim. Biophys. Acta 292, 256–270 (1973).
Cameron, L.M. & Hales, B.J. Investigation of CO binding and release from Mo-nitrogenase during catalytic turnover. Biochemistry 37, 9449–9456 (1998).
Spatzal, T., Perez, K.A., Einsle, O., Howard, J.B. & Rees, D.C. Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase. Science 345, 1620–1623 (2014).
Spatzal, T., Perez, K.A., Howard, J.B. & Rees, D.C. Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron-molybdenum cofactor. eLife 4, e11620 (2015).
Sippel, D. et al. Production and isolation of vanadium nitrogenase from Azotobacter vinelandii by molybdenum depletion. J. Biol. Inorg. Chem. 22, 161–168 (2017).
Kim, J. & Rees, D.C. Crystallographic structure and functional implications of the nitrogenase molybdenum-iron protein from Azotobacter vinelandii. Nature 360, 553–560 (1992).
Zhang, L. et al. The sixteenth iron in the nitrogenase MoFe protein. Angew. Chem. Int. Ed. Engl. 52, 10529–10532 (2013).
Dyer, D.H. et al. The three-dimensional structure of the core domain of Naf Y from Azotobacter vinelandii determined at 1.8-A resolution. J. Biol. Chem. 278, 32150–32156 (2003).
Homer, M.J., Paustian, T.D., Shah, V.K. & Roberts, G.P. The nifY product of Klebsiella pneumoniae is associated with apodinitrogenase and dissociates upon activation with the iron-molybdenum cofactor. J. Bacteriol. 175, 4907–4910 (1993).
Schmid, B. et al. Structure of a cofactor-deficient nitrogenase MoFe protein. Science 296, 352–356 (2002).
Hernandez, J.A. et al. A sterile α-motif domain in NafY targets apo-NifDK for iron-molybdenum cofactor delivery via a tethered domain. J. Biol. Chem. 286, 6321–6328 (2011).
Schindelin, H., Kisker, C., Schlessman, J.L., Howard, J.B. & Rees, D.C. Structure of ADP·AIF4--stabilized nitrogenase complex and its implications for signal transduction. Nature 387, 370–376 (1997).
Tezcan, F.A. et al. Nitrogenase complexes: multiple docking sites for a nucleotide switch protein. Science 309, 1377–1380 (2005).
Peters, J.W. et al. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36, 1181–1187 (1997).
Tittsworth, R.C. & Hales, B.J. Oxidative titration of the nitrogenase VFe protein from Azotobacter vinelandii: an example of redox-gated electron flow. Biochemistry 35, 479–487 (1996).
Rees, J.A. et al. The Fe-V cofactor of vanadium nitrogenase contains an interstitial carbon atom. Angew. Chem. Int. Ed. Engl. 54, 13249–13252 (2015).
Spatzal, T. et al. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940 (2011).
Kovacs, J.A. & Holm, R.H. Assembly of vanadium-iron-sulfur cubane clusters from mononuclear and linear trinuclear reactants. J. Am. Chem. Soc. 108, 340–341 (1986).
Varley, J.B., Wang, Y., Chan, K., Studt, F. & Nørskov, J.K. Mechanistic insights into nitrogen fixation by nitrogenase enzymes. Phys. Chem. Chem. Phys. 17, 29541–29547 (2015).
Setubal, J.C. et al. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J. Bacteriol. 191, 4534–4545 (2009).
Mendel, R.R. The molybdenum cofactor. J. Biol. Chem. 288, 13165–13172 (2013).
Lake, M.W., Wuebbens, M.M., Rajagopalan, K.V. & Schindelin, H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414, 325–329 (2001).
Wiig, J.A., Hu, Y., Lee, C.C. & Ribbe, M.W. Radical SAM-dependent carbon insertion into the nitrogenase M-cluster. Science 337, 1672–1675 (2012).
Lipscomb, J.D. Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol. 48, 371–399 (1994).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G.N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Acknowledgements
This work was supported by the European Research Council (grant no. 310656 to O.E.) and Deutsche Forschungsgemeinschaft (RTG 1976 and PP 1927 to O.E.) and the BIOSS Centre for Biological Signaling Studies at Albert-Ludwigs-Universität Freiburg (to O.E.). We thank the beamline staff at the Swiss Light Source, Villigen, Switzerland, G. Fritz and A. Brausemann for their excellent assistance with data collection, and S. Andrade for critical reading of the manuscript and helpful discussions.
Author information
Authors and Affiliations
Contributions
D.S. performed the experiments and built and refined the structural model; O.E. designed the experiments, built and refined the structural model and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–7 (PDF 2824 kb)
Rights and permissions
About this article
Cite this article
Sippel, D., Einsle, O. The structure of vanadium nitrogenase reveals an unusual bridging ligand. Nat Chem Biol 13, 956–960 (2017). https://doi.org/10.1038/nchembio.2428
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2428
This article is cited by
-
Structural insights into the iron nitrogenase complex
Nature Structural & Molecular Biology (2024)
-
Application and effectiveness of Methylobacterium symbioticum as a biological inoculant in maize and strawberry crops
Folia Microbiologica (2024)
-
Near ambient N2 fixation on solid electrodes versus enzymes and homogeneous catalysts
Nature Reviews Chemistry (2023)
-
Iron-only Fe-nitrogenase underscores common catalytic principles in biological nitrogen fixation
Nature Catalysis (2023)
-
Soil amendments for vanadium remediation: a review of remediation of vanadium in soil through chemical stabilization and bioremediation
Environmental Geochemistry and Health (2023)