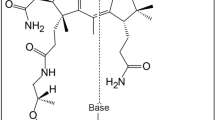
Abstract
Cobamides such as vitamin B12 are structurally conserved, cobalt-containing tetrapyrrole biomolecules that have essential biochemical functions in all domains of life. In organohalide respiration, a vital biological process for the global cycling of natural and anthropogenic organohalogens, cobamides are the requisite prosthetic groups for carbon–halogen bond-cleaving reductive dehalogenases. This study reports the biosynthesis of a new cobamide with unsubstituted purine as the lower base and assigns unsubstituted purine a biological function by demonstrating that Coα-purinyl-cobamide (purinyl-Cba) is the native prosthetic group in catalytically active tetrachloroethene reductive dehalogenases of Desulfitobacterium hafniense. Cobamides featuring different lower bases are not functionally equivalent, and purinyl-Cba elicits different physiological responses in corrinoid-auxotrophic, organohalide-respiring bacteria. Given that cobamide-dependent enzymes catalyze key steps in essential metabolic pathways, the discovery of a novel cobamide structure and the realization that lower bases can effectively modulate enzyme activities generate opportunities to manipulate functionalities of microbiomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
GenBank/EMBL/DDBJ
Protein Data Bank
References
Renz, P. in Chemistry and Biochemistry of B12 (ed. Banerjee, R.) 558–572 (John Wiley & Sons, Inc., 1999).
Gruber, K., Puffer, B. & Kräutler, B. Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids. Chem. Soc. Rev. 40, 4346–4363 (2011).
Kräutler, B. et al. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo-B12, a new type of a natural corrinoid. Helv. Chim. Acta 86, 3698–3716 (2003).
Im, J., Walshe-Langford, G.E., Moon, J.-W. & Löffler, F.E. Environmental fate of the next generation refrigerant 2,3,3,3-tetrafluoropropene (HFO-1234yf). Environ. Sci. Technol. 48, 13181–13187 (2014).
Maillard, J. et al. Characterization of the corrinoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl. Environ. Microbiol. 69, 4628–4638 (2003).
Liao, R.-Z., Chen, S.-L. & Siegbahn, P.E.M. Which oxidation state initiates dehalogenation in the B12-dependent enzyme NpRdhA: CoII, CoI, or Co0? ACS Catal. 5, 7350–7358 (2015).
Bommer, M. et al. Structural basis for organohalide respiration. Science 346, 455–458 (2014).
Payne, K.A.P. et al. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517, 513–516 (2015).
Löffler, F.E. et al. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635 (2013).
Yan, J. et al. The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi. ISME J. 10, 1092–1101 (2016).
Yi, S. et al. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl. Environ. Microbiol. 78, 7745–7752 (2012).
Mok, K.C. & Taga, M.E. Growth inhibition of Sporomusa ovata by incorporation of benzimidazole bases into cobamides. J. Bacteriol. 195, 1902–1911 (2013).
Stupperich, E., Eisinger, H.-J. & Kräutler, B. Identification of phenolyl cobamide from the homoacetogenic bacterium Sporomusa ovata. Eur. J. Biochem. 186, 657–661 (1989).
Villemur, R., Lanthier, M., Beaudet, R. & Lépine, F. The Desulfitobacterium genus. FEMS Microbiol. Rev. 30, 706–733 (2006).
Ding, C., Zhao, S. & He, J. A Desulfitobacterium sp. strain PR reductively dechlorinates both 1,1,1-trichloroethane and chloroform. Environ. Microbiol. 16, 3387–3397 (2014).
Suyama, A., Yamashita, M., Yoshino, S. & Furukawa, K. Molecular characterization of the PceA reductive dehalogenase of desulfitobacterium sp. strain Y51. J. Bacteriol. 184, 3419–3425 (2002).
Reinhold, A. et al. Impact of vitamin B12 on formation of the tetrachloroethene reductive dehalogenase in Desulfitobacterium hafniense strain Y51. Appl. Environ. Microbiol. 78, 8025–8032 (2012).
Nonaka, H. et al. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 188, 2262–2274 (2006).
Crofts, T.S., Seth, E.C., Hazra, A.B. & Taga, M.E. Cobamide structure depends on both lower ligand availability and CobT substrate specificity. Chem. Biol. 20, 1265–1274 (2013).
Yan, J., Im, J., Yang, Y. & Löffler, F.E. Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Phil. Trans. R. Soc. Lond. B 368, 20120320 (2013).
Keller, S. et al. Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ. Microbiol. 16, 3361–3369 (2014).
Guimarães, D.H., Weber, A., Klaiber, I., Vogler, B. & Renz, P. Guanylcobamide and hypoxanthylcobamide-corrinoids formed by Desulfovibrio vulgaris. Arch. Microbiol. 162, 272–276 (1994).
Stasyuk, O.A., Szatyłowicz, H. & Krygowski, T.M. Effect of the H-bonding on aromaticity of purine tautomers. J. Org. Chem. 77, 4035–4045 (2012).
Moffatt, B.A. & Ashihara, H. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book 1, e0018 (2002).
Kublik, A. et al. Identification of a multi-protein reductive dehalogenase complex in Dehalococcoides mccartyi strain CBDB1 suggests a protein-dependent respiratory electron transport chain obviating quinone involvement. Environ. Microbiol. 18, 3044–3056 (2016).
Dobbek, H., Svetlitchnyi, V., Gremer, L., Huber, R. & Meyer, O. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293, 1281–1285 (2001).
Iverson, T.M., Luna-Chavez, C., Cecchini, G. & Rees, D.C. Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284, 1961–1966 (1999).
Rosemeyer, H. The chemodiversity of purine as a constituent of natural products. Chem. Biodivers. 1, 361–401 (2004).
Löfgren, N. & Lüning, B. On the structure of nebularine. Acta Chem. Scand. 7, 225 (1953).
Nakamura, G. Studies on antibiotic actinomycetes. III. on Streptomyces producing 9-β-D-ribofuranosylpurine. J. Antibiot. 14, 94–97 (1961).
Cooper, R., Horan, A.C., Gunnarsson, I., Patel, M. & Truumees, I. Nebularine from a novel Microbispora sp. J. Ind. Microbiol. 1, 275–276 (1986).
Gordon, M.P. & Brown, G.B. A study of the metabolism of purine riboside. J. Biol. Chem. 220, 927–937 (1956).
Brown, E.G. & Konuk, M. Plant cytotoxicity of nebularine (purine riboside). Phytochemistry 37, 1589–1592 (1994).
el Kouni, M.H., Messier, N.J. & Cha, S. Treatment of schistosomiasis by purine nucleoside analogues in combination with nucleoside transport inhibitors. Biochem. Pharmacol. 36, 3815–3821 (1987).
Brown, E.G. & Konuk, M. Biosynthesis of nebularine (purine 9-β-D-ribofuranoside) involves enzymic release of hydroxylamine from adenosine. Phytochemistry 38, 61–71 (1995).
Ralevic, V. & Burnstock, G. Receptors for purines and pyrimidines. Pharmacol. Rev. 50, 413–492 (1998).
Burnstock, G. Purinergic signalling and disorders of the central nervous system. Nat. Rev. Drug Discov. 7, 575–590 (2008).
Di Virgilio, F. & Adinolfi, E. Extracellular purines, purinergic receptors and tumor growth. Oncogene 36, 293–303 (2017).
Miles, Z.D., McCarty, R.M., Molnar, G. & Bandarian, V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc. Natl. Acad. Sci. USA 108, 7368–7372 (2011).
Parks, J.M. et al. The genetic basis for bacterial mercury methylation. Science 339, 1332–1335 (2013).
Randaccio, L., Geremia, S., Demitri, N. & Wuerges, J. Vitamin B12: unique metalorganic compounds and the most complex vitamins. Molecules 15, 3228–3259 (2010).
Schneider, Z. in Comprehensive B12: Chemistry, Biochemistry, Nutrition, Ecology, Medicine (eds. Schneider, Z. & Stroiński, A.) 93–104 (Walter de Gruyter & Co., 1987).
Degnan, P.H., Taga, M.E. & Goodman, A.L. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778 (2014).
Suyama, A. et al. Isolation and characterization of Desulfitobacterium sp. strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Biosci. Biotechnol. Biochem. 65, 1474–1481 (2001).
Yan, J., Ritalahti, K.M., Wagner, D.D. & Löffler, F.E. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol. 78, 6630–6636 (2012).
Holliger, C. et al. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169, 313–321 (1998).
Hazra, A.B. et al. Anaerobic biosynthesis of the lower ligand of vitamin B12 . Proc. Natl. Acad. Sci. USA 112, 10792–10797 (2015).
Wang, P.-H. et al. Refined experimental annotation reveals conserved corrinoid autotrophy in chloroform-respiring Dehalobacter isolates. ISME J. 11, 626–640 (2017).
Tang, S. et al. Functional characterization of reductive dehalogenases by using blue native polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 79, 974–981 (2013).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Nesvizhskii, A.I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Ritalahti, K.M. et al. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72, 2765–2774 (2006).
Patiny, L. & Borel, A. ChemCalc: a building block for tomorrow's chemical infrastructure. J. Chem. Inf. Model. 53, 1223–1228 (2013).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Huang, L. et al. A family of metal-dependent phosphatases implicated in metabolite damage-control. Nat. Chem. Biol. 12, 621–627 (2016).
Hazra, A.B., Tran, J.L.A., Crofts, T.S. & Taga, M.E. Analysis of substrate specificity in CobT homologs reveals widespread preference for DMB, the lower axial ligand of vitamin (B12). Chem. Biol. 20, 1275–1285 (2013).
Acknowledgements
We thank J. Maillard, École Polytechnique Fédérale de Lausanne, France, for providing Dehalobacter restrictus strain PER-K23. We also thank R. Flick from the BioZone Mass Spectrometry facility, Toronto, for LC/MS assistance. This research was supported by a grant from the Superfund Research Program under the National Institute of Environmental Health Sciences (R01ES024294) to F.E.L., with additional support provided by the Strategic Environmental Research and Development Program (SERDP project ER-2312) to F.E.L. and by the Natural Science and Engineering Research Council of Canada (NSERC) Industrial Biocatalysis Network to E.A.E. Y. Yin acknowledges the financial support from the China-UT One-Hundred Scholars Program by the China Scholarship Council and the University of Tennessee.
Author information
Authors and Affiliations
Contributions
F.E.L., J.Y., and S.R.C. conceptualized the research and designed experiments. J.Y., M.B., B.S., Y. Yang, and Y. Yin performed cultivation work, corrinoid extraction and purification, and phylogenetic analyses. A.K.B. and A.T.F. performed LC–MS and structural analyses. P.W., O.M. and A.T.Q. performed BN–PAGE, enzyme assays, and proteomic analysis. N.J. generated cobT expression clones. All authors contributed to data analysis and interpretation, and J.Y., S.R.C., E.A.E., and F.E.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–5, Supplementary Figures 1–9 (PDF 2166 kb)
Rights and permissions
About this article
Cite this article
Yan, J., Bi, M., Bourdon, A. et al. Purinyl-cobamide is a native prosthetic group of reductive dehalogenases. Nat Chem Biol 14, 8–14 (2018). https://doi.org/10.1038/nchembio.2512
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2512