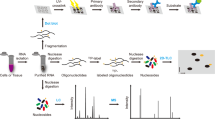
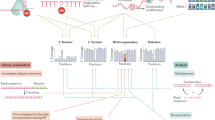
Abstract
Messenger RNA (mRNA) and long noncoding RNA (lncRNA) can be subjected to a variety of post-transcriptional modifications that markedly influence their fate and function. This concept of 'epitranscriptomic' modifications and the understanding of their function has been driven by new technologies for transcriptome-wide mapping of modified nucleotides using next-generation sequencing. Mapping technologies have successfully documented the location and prevalence of several modified nucleotides in the transcriptome. However, some mapping methods have led to proposals of pervasive novel RNA modifications that have subsequently been shown to be exceptionally rare. These controversies have resulted in confusion about the identity of the modified nucleotides comprising the epitranscriptome in mRNA and lncRNA. Here we discuss the different transcriptome-wide technologies for mapping modified nucleotides. We describe why these methods can have poor accuracy and specificity. Finally, we describe emerging strategies that minimize false positives and other pitfalls associated with mapping and measuring epitranscriptomic modifications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Meyer, K.D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012) The study in ref. 2 , along with that in ref. 1 , mapped a modified nucleotide (m6A) throughout mammalian transcriptomes for the first time, identifying specific transcripts containing modified residues and encouraging subsequent mapping of other modified residues.
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8, 284–296 (2014).
Meyer, K.D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Dominissini, D. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic (N1)-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Haussmann, I.U. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304 (2016).
Lence, T. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016).
Li, Z. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 (2017).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Batista, P.J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Hess, M.E. et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048 (2013).
Desrosiers, R., Friderici, K. & Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 71, 3971–3975 (1974).
Perry, R.P., Kelley, D.E., Friderici, K. & Rottman, F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell 4, 387–394 (1975).
Dubin, D.T. & Taylor, R.H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 2, 1653–1668 (1975).
Clancy, M.J., Shambaugh, M.E., Timpte, C.S. & Bokar, J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30, 4509–4518 (2002).
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008). The study in ref. 22 provides an analytical tool to selectively detect m6A in mRNA without contamination from rRNA or snRNA. This method takes advantage of the unique sequence context of m6A in mRNA.
Squires, J.E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Khoddami, V. & Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31, 458–464 (2013).
Hussain, S. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261 (2013) The studies in refs. 23,24,25 initially mapped m5C throughout the human transcriptome using diverse approaches.
Carlile, T.M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Lovejoy, A.F., Riordan, D.P. & Brown, P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014). This study in ref. 27 , along with those in refs. 26 and 8 , together initially mapped pseudouridine at nucleotide resolution in yeast and mammalian transcriptomes.
Delatte, B. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Dai, Q. et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017) The study in ref. 30 , which mapped m1A throughout the transcriptome at nucleotide resolution, demonstrated the importance of developing highly specific mapping approaches and employing rigorous validation measures in epitranscriptome mapping.
Gillen, A.E., Yamamoto, T.M., Kline, E., Hesselberth, J.R. & Kabos, P. Improvements to the HITS-CLIP protocol eliminate widespread mispriming artifacts. BMC Genomics 17, 338 (2016).
Legrand, C. et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 27, 1589–1596 (2017) The study in ref. 32 demonstrated the importance of establishing the noise level that occurs in an epitranscriptomic mapping method. The variability can be used to establish statistical criteria to determine if putatively mapped nucleotides are simply a product of statistical noise or if the nucleotide is statistically significant.
Gilbert, W.V., Bell, T.A. & Schaening, C. Messenger RNA modifications: form, distribution, and function. Science 352, 1408–1412 (2016).
Harcourt, E.M., Kietrys, A.M. & Kool, E.T. Chemical and structural effects of base modifications in messenger RNA. Nature 541, 339–346 (2017).
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Edelheit, S., Schwartz, S., Mumbach, M.R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602 (2013).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015) The study in ref. 38 employed antibody crosslinking to generate the first nucleotide-resolution map of m6A, the most prevalent modified residue in mRNA, and the first map of m6A m throughout mammalian transcriptomes. This study additionally demonstrates, using mapping, the extent of m6A antibodies' interaction with m6A m.
Munns, T.W., Oberst, R.J., Sims, H.F. & Liszewski, M.K. Antibody-nucleic acid complexes. Immunospecific recognition of 7-methylguanine- and N6-methyladenine-containing 5′-terminal oligonucleotides of mRNA. J. Biol. Chem. 254, 4327–4330 (1979).
Moss, B., Gershowitz, A., Weber, L.A. & Baglioni, C. Histone mRNAs contain blocked and methylated 5′ terminal sequences but lack methylated nucleosides at internal positions. Cell 10, 113–120 (1977).
Schibler, U. & Perry, R.P. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Res. 4, 4133–4149 (1977).
Ho, N.W. & Gilham, P.T. Reaction of pseudouridine and inosine with N-cyclohexyl-N′-β-(4-methylmorpholinium)ethylcarbodiimide. Biochemistry 10, 3651–3657 (1971).
Bakin, A. & Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762 (1993).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013) The study in ref. 45 provides an analytical method for biochemically detecting and quantifying modified residues at any nucleotide in any transcript of interest. Although this method was originally described for m6A, it can easily be adapted to validate the presence of nearly any modified nucleotide of interest.
Liu, Y. & Santi, D.V. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl. Acad. Sci. USA 97, 8263–8265 (2000).
Hauenschild, R. et al. The reverse transcription signature of N-1-methyladenosine in RNA-seq is sequence dependent. Nucleic Acids Res. 43, 9950–9964 (2015).
Nottingham, R.M. et al. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22, 597–613 (2016).
Macon, J.B. & Wolfenden, R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry 7, 3453–3458 (1968).
Wu, Z., Wang, X. & Zhang, X. Using non-uniform read distribution models to improve isoform expression inference in RNA-seq. Bioinformatics 27, 502–508 (2011).
Li, X. et al. Base-Resolution Mapping Reveals Distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e (2017).
Machnicka, M.A. et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Furuichi, Y. et al. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl. Acad. Sci. USA 72, 1904–1908 (1975).
Amort, T. et al. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 10, 1003–1008 (2013).
Schaefer, M., Pollex, T., Hanna, K. & Lyko, F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12 (2009).
Safra, M., Nir, R., Farouq, D., Vainberg Slutskin, I. & Schwartz, S. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 27, 393–406 (2017).
Liu, L. et al. Decomposition of RNA methylome reveals co-methylation patterns induced by latent enzymatic regulators of the epitranscriptome. Mol. Biosyst. 11, 262–274 (2015).
Mokry, M. et al. Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PLoS One 5, e15092 (2010).
Levin, J.Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 (2010).
Ke, S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017).
Wei, C., Gershowitz, A. & Moss, B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253 (1975).
Kruse, S. et al. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci. Rep. 1, 126 (2011). The study in ref. 62 provides an analytical method for biochemically detecting and quantifying modifications of the mRNA cap.
Molinie, B. et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692–698 (2016).
Auffray, C. & Rougeon, F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur. J. Biochem. 107, 303–314 (1980).
Zhao, W. et al. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics 15, 419 (2014).
Sultan, M. et al. Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC Genomics 15, 675 (2014).
Cui, P. et al. A comparison between ribo-minus RNA-sequencing and polyA-selected RNA-sequencing. Genomics 96, 259–265 (2010).
Shimba, S., Bokar, J.A., Rottman, F. & Reddy, R. Accurate and efficient N6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 23, 2421–2426 (1995).
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Śledź, P. & Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 5, e18434 (2016).
Wang, P., Doxtader, K.A. & Nam, Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016).
Wang, X. et al. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 (2016).
Van Gelder, R.N. et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. USA 87, 1663–1667 (1990).
Baugh, L.R., Hill, A.A., Brown, E.L. & Hunter, C.P. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 29, E29 (2001).
Olarerin-George, A.O. & Jaffrey, S.R. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics 33, 1563–1564 (2017).
Bailey, T.L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Acknowledgements
We thank all members of the Jaffrey lab for helpful comments and suggestions. This work was supported by NIH grants R01DA037755 (S.R.J.) and UL1 TR000457 (A.G.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Grozhik, A., Jaffrey, S. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat Chem Biol 14, 215–225 (2018). https://doi.org/10.1038/nchembio.2546
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2546
This article is cited by
-
Navigating the pitfalls of mapping DNA and RNA modifications
Nature Reviews Genetics (2023)
-
MePMe-seq: antibody-free simultaneous m6A and m5C mapping in mRNA by metabolic propargyl labeling and sequencing
Nature Communications (2023)
-
Epitranscriptomic subtyping, visualization, and denoising by global motif visualization
Nature Communications (2023)
-
Detection of m6A from direct RNA sequencing using a multiple instance learning framework
Nature Methods (2022)
-
Developmental mRNA m5C landscape and regulatory innovations of massive m5C modification of maternal mRNAs in animals
Nature Communications (2022)