Abstract
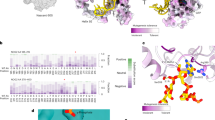
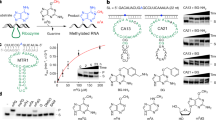
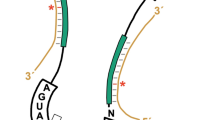
Methylation of nucleotides in ribosomal RNAs (rRNAs) is a ubiquitous feature that occurs in all living organisms. Identification of all enzymes responsible for rRNA methylation, as well as mapping of all modified rRNA residues, is now complete for a number of model species, such as Escherichia coli and Saccharomyces cerevisiae. Recent high-resolution structures of bacterial ribosomes provided the first direct visualization of methylated nucleotides. The structures of ribosomes from various organisms and organelles have also lately become available, enabling comparative structure-based analysis of rRNA methylation sites in various taxonomic groups. In addition to the conserved core of modified residues in ribosomes from the majority of studied organisms, structural analysis points to the functional roles of some of the rRNA methylations, which are discussed in this Review in an evolutionary context.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Machnicka, M.A. et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 41, D262–D267 (2013). Ref. 1 contains an update to the comprehensive database of known types of RNA modifications, the chemical structures of modified ribonucleosides, their biosynthetic pathways, RNA-modifying enzymes and the location of modified residues in RNA sequences.
Seistrup, K.H. et al. Bypassing rRNA methylation by RsmA/Dim1during ribosome maturation in the hyperthermophilic archaeon Nanoarchaeum equitans. Nucleic Acids Res. 45, 2007–2015 (2017).
Piekna-Przybylska, D., Decatur, W.A. & Fournier, M.J. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 36, D178–D183 (2008).
Fu, Y., Dominissini, D., Rechavi, G. & He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014).
Meyer, K.D. & Jaffrey, S.R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Golovina, A.Y. et al. The last rRNA methyltransferase of E. coli revealed: the yhiR gene encodes adenine-N6 methyltransferase specific for modification of A2030 of 23S ribosomal RNA. RNA 18, 1725–1734 (2012). With the work in ref. 6 , the list of E. coli rRNA methyltransferases is completed. It allows conducting a comprehensive analysis of the entire rRNA methylation system in the most studied model organism.
Sharma, S. & Lafontaine, D.L. 'View from a bridge': a new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 40, 560–575 (2015).
Liu, M. & Douthwaite, S. Methylation at nucleotide G745 or G748 in 23S rRNA distinguishes Gram-negative from Gram-positive bacteria. Mol. Microbiol. 44, 195–204 (2002).
Desmolaize, B. et al. A single methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939 modifications in Bacillus subtilis 23S rRNA. Nucleic Acids Res. 39, 9368–9375 (2011).
Lartigue, C. et al. The flavoprotein Mcap0476 (RlmFO) catalyzes m5U1939 modification in Mycoplasma capricolum 23S rRNA. Nucleic Acids Res. 42, 8073–8082 (2014). Refs. 9 and 10 describe examples of the divergent and convergent evolution of rRNA methyltransferases, respectively.
Guymon, R., Pomerantz, S.C., Crain, P.F. & McCloskey, J.A. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry 45, 4888–4899 (2006).
Schosserer, M. et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 6, 6158 (2015).
Wurm, J.P. et al. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 38, 2387–2398 (2010).
Meyer, B. et al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Ψ1191 in yeast 18S rRNA. Nucleic Acids Res. 39, 1526–1537 (2011).
Meyer, B. et al. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res. 44, 4304–4316 (2016).
Zorbas, C. et al. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell 26, 2080–2095 (2015).
Waku, T. et al. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J. Cell Sci. 129, 2382–2393 (2016).
Polikanov, Y.S., Melnikov, S.V., Söll, D. & Steitz, T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015).
Noeske, J. et al. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 (2015). These studies describe structures of the bacterial 70S ribosomes either from T. thermophilus (ref. 18 ) or E. coli (ref. 19 ) that allowed, for the first time, the visualization of every individual nucleotide modification in the bacterial ribosomes at the highest resolution reported to date.
Ben-Shem, A. et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 (2011). The study in ref. 20 reports the first crystal structure of the 80S ribosome from yeast S. cerevisiae.
Khatter, H., Myasnikov, A.G., Natchiar, S.K. & Klaholz, B.P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015). The study in ref. 21 reports the highest-resolution cryo-EM structure of the 80S ribosome from human.
Anger, A.M. et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013). The study in ref. 22 reports the first cryo-EM structure of the 80S ribosome from human.
Amunts, A., Brown, A., Toots, J., Scheres, S.H.W. & Ramakrishnan, V. Ribosome. The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015).
Desai, N., Brown, A., Amunts, A. & Ramakrishnan, V. The structure of the yeast mitochondrial ribosome. Science 355, 528–531 (2017). The first two cryo-EM structures of the complete ribosomes from human and yeast mitochondria are reported in refs. 23 and 24, respectively.
Sergiev, P.V. et al. in Ribosomes (eds. Rodnina, M.V., Wintermeyer, W. & Green, R.) 97–110 (Springer Vienna, 2011).
Siibak, T. & Remme, J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA 16, 2023–2032 (2010).
Kiss-László, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M. & Kiss, T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85, 1077–1088 (1996).
Ni, J., Tien, A.L. & Fournier, M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89, 565–573 (1997).
Yang, J. et al. Mapping of complete set of ribose and base modifications of yeast rRNA by RP-HPLC and mung bean nuclease assay. PLoS One 11, e0168873 (2016) Ref. 29 completes mapping of all modified residues within yeast rRNA.
Piekna-Przybylska, D., Decatur, W.A. & Fournier, M.J. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA 13, 305–312 (2007).
Lestrade, L. & Weber, M.J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 34, D158–D162 (2006).
Kirpekar, F., Hansen, L.H., Rasmussen, A., Poehlsgaard, J. & Vester, B. The archaeon Haloarcula marismortui has few modifications in the central parts of its 23S ribosomal RNA. J. Mol. Biol. 348, 563–573 (2005).
Sloan, K.E. et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152 (2017).
Demirci, H. et al. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA 16, 2319–2324 (2010).
Baudin-Baillieu, A. et al. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 37, 7665–7677 (2009).
Prokhorova, I.V. et al. Modified nucleotides m2G966/m5C967 of Escherichia coli 16S rRNA are required for attenuation of tryptophan operon. Sci. Rep. 3, 3236 (2013).
van Buul, C.P., Visser, W. & van Knippenberg, P.H. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett. 177, 119–124 (1984).
Okamoto, S. et al. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 63, 1096–1106 (2007).
Popova, A.M. & Williamson, J.R. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J. Am. Chem. Soc. 136, 2058–2069 (2014). Ref. 39 has evidence for the temporal pattern of rRNA modifications that occur during the assembly of ribosomal subunits.
Birkedal, U. et al. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Edn Engl. 54, 451–455 (2015).
Kornprobst, M. et al. Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166, 380–393 (2016).
Sloan, K.E. et al. The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res. 43, 553–564 (2015).
Lapeyre, B. & Purushothaman, S.K. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell 16, 663–669 (2004).
Létoquart, J. et al. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. USA 111, E5518–E5526 (2014).
Maden, E.H. & Wakeman, J.A. Pseudouridine distribution in mammalian 18S ribosomal RNA. A major cluster in the central region of the molecule. Biochem. J. 249, 459–464 (1988).
Samarsky, D.A., Balakin, A.G. & Fournier, M.J. Characterization of three new snRNAs from Saccharomyces cerevisiae: snR34, snR35 and snR36. Nucleic Acids Res. 23, 2548–2554 (1995).
Arenz, S. & Wilson, D.N. Blast from the past: reassessing forgotten translation inhibitors, antibiotic selectivity, and resistance mechanisms to aid drug development. Mol. Cell 61, 3–14 (2016).
Armistead, J. et al. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am. J. Hum. Genet. 84, 728–739 (2009).
Oie, S. et al. Hepatic rRNA transcription regulates high-fat-diet-induced obesity. Cell Rep. 7, 807–820 (2014).
Koeck, T. et al. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab. 13, 80–91 (2011).
Bourgeois, G. et al. Eukaryotic rRNA modification by yeast 5-methylcytosine-methyltransferases and human proliferation-associated antigen p120. PLoS One 10, e0133321 (2015).
Poldermans, B., Roza, L. & Van Knippenberg, P.H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16S ribosomal RNA of Escherichia coli. III. Purification and properties of the methylating enzyme and methylase-30S interactions. J. Biol. Chem. 254, 9094–9100 (1979).
Lafontaine, D., Delcour, J., Glasser, A.L., Desgrès, J. & Vandenhaute, J. The DIM1 gene responsible for the conserved m62Am62A dimethylation in the 3′-terminal loop of 18S rRNA is essential in yeast. J. Mol. Biol. 241, 492–497 (1994).
Seidel-Rogol, B.L., McCulloch, V. & Shadel, G.S. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 33, 23–24 (2003).
Cotney, J., McKay, S.E. & Shadel, G.S. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum. Mol. Genet. 18, 2670–2682 (2009).
Connolly, K., Rife, J.P. & Culver, G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 70, 1062–1075 (2008). The study in ref. 56 reports the first evidence of the second function of KsgA methyltransferase as ribosome assembly factor.
Lafontaine, D., Vandenhaute, J. & Tollervey, D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 9, 2470–2481 (1995).
White, J. et al. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell. Biol. 28, 3151–3161 (2008).
Haag, S., Kretschmer, J. & Bohnsack, M.T. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 21, 180–187 (2015).
Lövgren, J.M. & Wikström, P.M. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J. Bacteriol. 183, 6957–6960 (2001).
Caldas, T. et al. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem. 275, 16414–16419 (2000).
Tan, J., Jakob, U. & Bardwell, J.C. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184, 2692–2698 (2002).
Pintard, L., Bujnicki, J.M., Lapeyre, B. & Bonnerot, C. MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 21, 1139–1147 (2002).
Lesnyak, D.V. et al. Methyltransferase that modifies guanine 966 of the 16S rRNA: functional identification and tertiary structure. J. Biol. Chem. 282, 5880–5887 (2007).
Tscherne, J.S. et al. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry 38, 1884–1892 (1999).
Arora, S. et al. Distinctive contributions of the ribosomal P-site elements m2G966, m5C967 and the C-terminal tail of the S9 protein in the fidelity of initiation of translation in Escherichia coli. Nucleic Acids Res. 41, 4963–4975 (2013).
Burakovsky, D.E. et al. Impact of methylations of m2G966/m5C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res. 40, 7885–7895 (2012).
Kimura, S. & Suzuki, T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 38, 1341–1352 (2010).
Toh, S.M., Xiong, L., Bae, T. & Mankin, A.S. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA 14, 98–106 (2008).
Giessing, A.M. et al. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15, 327–336 (2009).
Polikanov, Y.S. et al. Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics Hygromycin A and A201A. Mol. Cell 58, 832–844 (2015).
Kimura, S., Sakai, Y., Ishiguro, K. & Suzuki, T. Biogenesis and iron-dependency of ribosomal RNA hydroxylation. Nucleic Acids Res. 45, 12974–12986 (2017).
Maden, B.E. Locations of methyl groups in 28S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J. Mol. Biol. 201, 289–314 (1988).
Raychaudhuri, S., Conrad, J., Hall, B.G. & Ofengand, J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4, 1407–1417 (1998).
Urbonavicius, J., Skouloubris, S., Myllykallio, H. & Grosjean, H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria--evolutionary implications. Nucleic Acids Res. 33, 3955–3964 (2005).
Madsen, C.T., Mengel-Jørgensen, J., Kirpekar, F. & Douthwaite, S. Identifying the methyltransferases for m5U747 and m5U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 31, 4738–4746 (2003).
Chujo, T. & Suzuki, T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 18, 2269–2276 (2012).
Bar-Yaacov, D. et al. Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol. 14, e1002557 (2016).
Balakin, A.G., Schneider, G.S., Corbett, M.S., Ni, J. & Fournier, M.J. SnR31, snR32, and snR33: three novel, non-essential snRNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 21, 5391–5397 (1993).
Tscherne, J.S., Nurse, K., Popienick, P. & Ofengand, J. Purification, cloning, and characterization of the 16S RNA m2G1207 methyltransferase from Escherichia coli. J. Biol. Chem. 274, 924–929 (1999).
Lowe, T.M. & Eddy, S.R. A computational screen for methylation guide snoRNAs in yeast. Science 283, 1168–1171 (1999).
Metodiev, M.D. et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 10, e1004110 (2014).
Andersen, N.M. & Douthwaite, S. YebU is a m5C methyltransferase specific for 16S rRNA nucleotide 1407. J. Mol. Biol. 359, 777–786 (2006).
Basturea, G.N., Rudd, K.E. & Deutscher, M.P. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA 12, 426–434 (2006).
Basturea, G.N., Dague, D.R., Deutscher, M.P. & Rudd, K.E. YhiQ is RsmJ, the methyltransferase responsible for methylation of G1516 in 16S rRNA of E. coli. J. Mol. Biol. 415, 16–21 (2012).
Peifer, C. et al. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 41, 1151–1163 (2013).
Gustafsson, C. & Persson, B.C. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 180, 359–365 (1998).
Sergiev, P.V., Serebryakova, M.V., Bogdanov, A.A. & Dontsova, O.A. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol. 375, 291–300 (2008).
Sharma, S., Watzinger, P., Kötter, P. & Entian, K.D. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 41, 5428–5443 (2013).
Sergiev, P.V., Lesnyak, D.V., Bogdanov, A.A. & Dontsova, O.A. Identification of Escherichia coli m2G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23S rRNA. J. Mol. Biol. 364, 26–31 (2006).
Purta, E., Kaminska, K.H., Kasprzak, J.M., Bujnicki, J.M. & Douthwaite, S. YbeA is the m3Psi methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA 14, 2234–2244 (2008).
Badis, G., Fromont-Racine, M. & Jacquier, A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 9, 771–779 (2003).
Ganot, P., Bortolin, M.L. & Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89, 799–809 (1997).
Sharma, S., Yang, J., Watzinger, P., Kötter, P. & Entian, K.D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 41, 9062–9076 (2013).
Purta, E., O'Connor, M., Bujnicki, J.M. & Douthwaite, S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J. Mol. Biol. 383, 641–651 (2008).
Kimura, S. et al. Base methylations in the double-stranded RNA by a fused methyltransferase bearing unwinding activity. Nucleic Acids Res. 40, 4071–4085 (2012). Ref. 96 reports identification of the first rRNA-methyltransferase with two catalytic domains, each targeting a specific nucleotide.
Lee, K.W. & Bogenhagen, D.F. Assignment of 2′-O-methyltransferases to modification sites on the mammalian mitochondrial large subunit 16S ribosomal RNA (rRNA). J. Biol. Chem. 289, 24936–24942 (2014).
Sharma, S. et al. Identification of novel methyltransferases, Bmt5 and Bmt6, responsible for the m3U methylations of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 42, 3246–3260 (2014).
Lesnyak, D.V., Sergiev, P.V., Bogdanov, A.A. & Dontsova, O.A. Identification of Escherichia coli m2G methyltransferases: I. the ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J. Mol. Biol. 364, 20–25 (2006).
Purta, E., O'Connor, M., Bujnicki, J.M. & Douthwaite, S. S. YgdE is the 2′-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol. 72, 1147–1158 (2009).
Acknowledgements
We thank P. Moore, M. Gagnon, J. Brown, and M. Svetlov for critical reading of the manuscript and valuable suggestions. We also thank all members of the P.V.S. and Y.S.P. laboratories for discussions and critical feedback. This work was supported by Illinois State startup funds (to Y.S.P.); Russian Foundation for Basic Research (16-04-01100 to P.V.S.); Russian Science Foundation (14-14-00072 to P.V.S.); and Moscow University Development Program (PNR 5.13 to P.V.S.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Sergiev, P., Aleksashin, N., Chugunova, A. et al. Structural and evolutionary insights into ribosomal RNA methylation. Nat Chem Biol 14, 226–235 (2018). https://doi.org/10.1038/nchembio.2569
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2569
This article is cited by
-
N6-methyladenosine modification—a key player in viral infection
Cellular & Molecular Biology Letters (2023)
-
Navigating the pitfalls of mapping DNA and RNA modifications
Nature Reviews Genetics (2023)
-
MePMe-seq: antibody-free simultaneous m6A and m5C mapping in mRNA by metabolic propargyl labeling and sequencing
Nature Communications (2023)
-
Biological roles of adenine methylation in RNA
Nature Reviews Genetics (2023)
-
Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nature Communications (2022)