Abstract
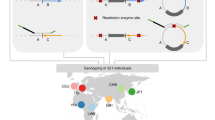
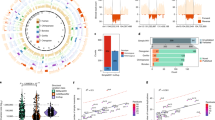
The 17q21.31 inversion polymorphism exists either as direct (H1) or inverted (H2) haplotypes with differential predispositions to disease and selection. We investigated its genetic diversity in 2,700 individuals, with an emphasis on African populations. We characterize eight structural haplotypes due to complex rearrangements that vary in size from 1.08–1.49 Mb and provide evidence for a 30-kb H1-H2 double recombination event. We show that recurrent partial duplications of the KANSL1 gene have occurred on both the H1 and H2 haplotypes and have risen to high frequency in European populations. We identify a likely ancestral H2 haplotype (H2′) lacking these duplications that is enriched among African hunter-gatherer groups yet essentially absent from West African populations. Whereas H1 and H2 segmental duplications arose independently and before human migration out of Africa, they have reached high frequencies recently among Europeans, either because of extraordinary genetic drift or selective sweeps.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
11 July 2012
In the version of this article initially published online, the final sentence in the second paragraph of the Results section incorrectly referred to the H2-specific duplication as CNP205. The correct designation for the H2-specific duplication is CNP155. Also, in the legend to Figure 5, the phrases describing panels c and d were inadvertently switched. These errors have been corrected for the print, PDF and HTML versions of this article.
References
Dobzhansky, T. The genetics of natural populations. Genetics 35, 288–302 (1950).
Dobzhansky, T. & Sturtevant, A.H. Inversions in the chromosomes of Drosophila pseudoobscura. Genetics 23, 28–64 (1938).
Lowry, D.B. & Willis, J.H. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8, e1000500 (2010).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (2005).
Kidd, J.M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
Bailey, J.A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002).
Sharp, A.J. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum. Mutat. 30, 135–144 (2009).
Lupski, J.R. Genome structural variation and sporadic disease traits. Nat. Genet. 38, 974–976 (2006).
Antonacci, F. et al. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet. 18, 2555–2566 (2009).
Stefansson, H. et al. A common inversion under selection in Europeans. Nat. Genet. 37, 129–137 (2005).
Sharp, A.J. et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 38, 1038–1042 (2006).
Koolen, D.A. et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 38, 999–1001 (2006).
Zody, M.C. et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 40, 1076–1083 (2008).
Koolen, D.A. et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J. Med. Genet. 45, 710–720 (2008).
Sudmant, P.H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Donnelly, M.P. et al. The distribution and most recent common ancestor of the 17q21 inversion in humans. Am. J. Hum. Genet. 86, 161–171 (2010).
Schuster, S.C. et al. Complete Khoisan and Bantu genomes from southern Africa. Nature 463, 943–947 (2010).
Baker, M. et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum. Mol. Genet. 8, 711–715 (1999).
Rao, P.N., Li, W., Vissers, L.E., Veltman, J.A. & Ophoff, R.A. Recurrent inversion events at 17q21.31 microdeletion locus are linked to the MAPT H2 haplotype. Cytogenet. Genome Res. 129, 275–279 (2010).
Boettger, L.M., Handsaker, R.E., Zody, M.C. & McCarroll, S.A. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat. Genet. published online, doi:10.1038/ng.2334 (1 July 2012).
Henn, B.M. et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. USA 108, 5154–5162 (2011).
Kidd, J.M. et al. Haplotype sorting using human fosmid clone end-sequence pairs. Genome Res. 18, 2016–2023 (2008).
Cargill, M. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22, 231–238 (1999).
Heim, C. et al. Effect of childhood trauma on adult depression and neuroendocrine function: sex-specific moderation by CRH receptor 1 gene. Front. Behav. Neurosci. 3, 41 (2009).
Liu, Z. et al. Association of corticotropin-releasing hormone receptor 1 gene SNP and haplotype with major depression. Neurosci. Lett. 404, 358–362 (2006).
Liu, Z. et al. Association study of corticotropin-releasing hormone receptor 1 gene polymorphisms and antidepressant response in major depressive disorders. Neurosci. Lett. 414, 155–158 (2007).
Bradley, R.G. et al. Influence of child abuse on adult depression: moderation by the corticotropin-releasing hormone receptor gene. Arch. Gen. Psychiatry 65, 190–200 (2008).
Polanczyk, G. et al. Protective effect of CRHR1 gene variants on the development of adult depression following childhood maltreatment: replication and extension. Arch. Gen. Psychiatry 66, 978–985 (2009).
Shaw-Smith, C. et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 38, 1032–1037 (2006).
Cooper, G.M. et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846 (2011).
Mefford, H.C. et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 19, 1579–1585 (2009).
Tishkoff, S.A. et al. The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (2009).
Fledel-Alon, A. et al. Variation in human recombination rates and its genetic determinants. PLoS ONE 6, e20321 (2011).
Kong, A. et al. Recombination rate and reproductive success in humans. Nat. Genet. 36, 1203–1206 (2004).
Coop, G., Wen, X., Ober, C., Pritchard, J.K. & Przeworski, M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319, 1395–1398 (2008).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
Simón-Sánchez, J. et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 41, 1308–1312 (2009).
Koolen, D.A. et al. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat. Genet. 44, 639–641 (2012).
Zollino, M. et al. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat. Genet. 44, 636–638 (2012).
Tobin, J.E. et al. Haplotypes and gene expression implicate the MAPT region for Parkinson disease: the GenePD Study. Neurology 71, 28–34 (2008).
Skipper, L. et al. Linkage disequilibrium and association of MAPT H1 in Parkinson disease. Am. J. Hum. Genet. 75, 669–677 (2004).
Myers, A.J. et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol. Dis. 25, 561–570 (2007).
Pittman, A.M. et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum. Mol. Genet. 13, 1267–1274 (2004).
Höglinger, G.U. et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat. Genet. 43, 699–705 (2011).
Setó-Salvia, N. et al. Dementia risk in Parkinson disease: disentangling the role of MAPT haplotypes. Arch. Neurol. 68, 359–364 (2011).
Dubourg, C. et al. Clinical and molecular characterization of 17q21.31 microdeletion syndrome in 14 French patients with mental retardation. Eur. J. Med. Genet. 54, 144–151 (2011).
Dawkins, R. et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol. Rev. 167, 275–304 (1999).
Reich, D. et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010).
Green, R.E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010).
Jiang, Z. et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 39, 1361–1368 (2007).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Antonacci, F. et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat. Genet. 42, 745–750 (2010).
Adey, A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 11, R119 (2010).
Igartua, C. et al. Targeted enrichment of specific regions in the human genome by array hybridization. Curr. Protoc. Hum. Genet. Chapter 18, Unit 18 3 (2010).
Campbell, C.D. et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am. J. Hum. Genet. 88, 317–332 (2011).
Browning, B.L. & Browning, S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009).
Browning, S.R. & Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Felsenstein, J. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166 (1989).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Acknowledgements
We thank J. Akey, M. Dennis and B. Dumont for helpful discussions and C. Alkan for computational assistance. We thank Z. Jiang for his initial work on the H1-H2 alignments. We are grateful to T. Brown for assistance with manuscript preparation, to C. Lee for technical assistance and to the anonymous reviewers of this paper who provided insightful comments. We thank the 1000 Genomes Project Consortium for access to unpublished sequence data for the 17q21.31 locus. K.M.S. was supported by a Ruth L. Kirschstein National Research Service Award (NRSA) training grant to the University of Washington (T32HG00035) and an individual NRSA Fellowship (F32GM097807). C.D.C. was supported by an individual NRSA Fellowship (F32HG006070). P.H.S. was supported by a Natural Sciences and Engineering Research Council of Canada Fellowship. J.M.K. was supported by a Ruth L. Kirschstein NRSA training grant to Stanford University (T32HG000044). This work was supported by the US National Institutes of Health (grants HG002385 and HG004120 to E.E.E.). E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
K.M.S., F.A. and E.E.E. designed the study. K.M.S. performed aCGH, genotyping and sequence analysis. F.A. performed FISH experiments and fosmid shotgun sequencing library construction. P.H.S. performed read depth–based copy-number analysis. J.M.K. performed sequence analysis on the double recombination region. C.D.C. performed aCGH analysis. L.V. and M.M. performed whole-genome shotgun sequencing library construction and PCR genotyping. L.S. and W.B. performed PCR genotyping and SNP array genotyping. M.I., G.L., T.B.N., S.A.O., J.-M.B. and A.F. contributed to African sample collection. M.P.D. and K.K.K. contributed to H2 Diversity Panel sample collection and genotyping. S.A.T. contributed to African sample collection and SNP array data. K.M.S., F.A., J.M.K., S.A.T. and E.E.E. contributed to data interpretation. K.M.S., F.A. and E.E.E. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.E.E. is on the scientific advisory boards for Pacific Biosciences, Inc., SynapDx Corp and DNAnexus, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–4 and Supplementary Tables 1 and 3–11 (PDF 1517 kb)
Supplementary Table 2
Individual-level 17q21.31 haplotype results (XLS 627 kb)
Rights and permissions
About this article
Cite this article
Steinberg, K., Antonacci, F., Sudmant, P. et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet 44, 872–880 (2012). https://doi.org/10.1038/ng.2335
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2335
This article is cited by
-
A new blood DNA methylation signature for Koolen-de Vries syndrome: Classification of missense KANSL1 variants and comparison to fibroblast cells
European Journal of Human Genetics (2024)
-
Neolithic expansion and the 17q21.31 inversion in Iberia: an evolutionary approach to H2 haplotype distribution in the Near East and Europe
Molecular Genetics and Genomics (2023)
-
Genome-wide profiling of 5-hydroxymethylcytosines in circulating cell-free DNA reveals population-specific pathways in the development of multiple myeloma
Journal of Hematology & Oncology (2022)
-
Association studies between chromosomal regions 1q21.3, 5q21.3, 14q21.2 and 17q21.31 and numbers of children in Poland
Scientific Reports (2022)
-
The early-life exposome modulates the effect of polymorphic inversions on DNA methylation
Communications Biology (2022)