Abstract
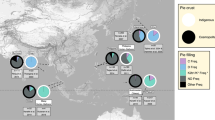
Tuberculosis caused 20% of all human deaths in the Western world between the seventeenth and nineteenth centuries and remains a cause of high mortality in developing countries. In analogy to other crowd diseases, the origin of human tuberculosis has been associated with the Neolithic Demographic Transition, but recent studies point to a much earlier origin. We analyzed the whole genomes of 259 M. tuberculosis complex (MTBC) strains and used this data set to characterize global diversity and to reconstruct the evolutionary history of this pathogen. Coalescent analyses indicate that MTBC emerged about 70,000 years ago, accompanied migrations of anatomically modern humans out of Africa and expanded as a consequence of increases in human population density during the Neolithic period. This long coevolutionary history is consistent with MTBC displaying characteristics indicative of adaptation to both low and high host densities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Sequence Read Archive
References
Wilson, L.G. Commentary: medicine, population, and tuberculosis. Int. J. Epidemiol. 34, 521–524 (2005).
World Health Organization. The Global Plan to STOP TB 2011–2015 (World Health Organization, Geneva, 2011).
Wolfe, N.D., Dunavan, C.P. & Diamond, J. Origins of major human infectious diseases. Nature 447, 279–283 (2007).
Diamond, J. Guns, Germs, and Steel: The Fates of Human Societies 496 (W.W. Norton & Company, New York, 1999).
Blaser, M.J. & Kirschner, D. The equilibria that allow bacterial persistence in human hosts. Nature 449, 843–849 (2007).
Berg, G. The prognosis of open pulmonary tuberculosis: a clinical-statistical analysis. J. Am. Med. Assoc. 114, 1954–1955 (1940).
Barry, C.E. III et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855 (2009).
Hershberg, R. et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6, e311 (2008).
Mostowy, S., Cousins, D., Brinkman, J., Aranaz, A. & Behr, M.A. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186, 74–80 (2002).
Wirth, T. et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4, e1000160 (2008).
Brosch, R. et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99, 3684–3689 (2002).
Gutierrez, M.C. et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 1, e5 (2005).
Gagneux, S. et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103, 2869–2873 (2006).
Firdessa, R. et al. Mycobacterial lineages causing pulmonary and extrapulmonary tuberculosis in Ethiopia. Emerg. Infect. Dis. 19, 460–463 (2013).
Namouchi, A., Didelot, X., Schöck, U., Gicquel, B. & Rocha, E.P.C. After the bottleneck: genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 22, 721–734 (2012).
Hirsh, A.E., Tsolaki, A.G., DeRiemer, K., Feldman, M.W. & Small, P.M. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101, 4871–4876 (2004).
Comas, I. & Gagneux, S. The past and future of tuberculosis research. PLoS Pathog. 5, e1000600 (2009).
Behar, D.M. et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012).
Drummond, A.J., Ho, S.Y.W., Phillips, M.J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (2006).
Bos, K.I. et al. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478, 506–510 (2011).
Mutreja, A. et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465 (2011).
Morelli, G. et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42, 1140–1143 (2010).
Djelouadji, Z., Raoult, D. & Drancourt, M. Palaeogenomics of Mycobacterium tuberculosis: epidemic bursts with a degrading genome. Lancet Infect. Dis. 11, 641–650 (2011).
Ford, C.B. et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat. Genet. 43, 482–486 (2011).
Walker, T.M. et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146 (2013).
Morelli, G. et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 6, e1001036 (2010).
Ho, S.Y.W. et al. Time-dependent rates of molecular evolution. Mol. Ecol. 20, 3087–3101 (2011).
Holt, K.E. et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat. Genet. 44, 1056–1059 (2012).
Croucher, N.J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011).
Harris, S.R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2010).
Soares, P. et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 84, 740–759 (2009).
Soares, P. et al. The expansion of mtDNA haplogroup L3 within and out of Africa. Mol. Biol. Evol. 29, 915–927 (2012).
Rasmussen, M. et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science 334, 94–98 (2011).
Henn, B.M., Cavalli-Sforza, L.L. & Feldman, M.W. The great human expansion. Proc. Natl. Acad. Sci. USA 109, 17758–17764 (2012).
Stewart, J.R. & Stringer, C.B. Human evolution out of Africa: the role of refugia and climate change. Science 335, 1317–1321 (2012).
Jin, L. & Su, B. Natives or immigrant: modern human origins in East Asia. Nat. Rev. Genet. 1, 126–133 (2000).
Bellwood, P. & Oxenham, M. The Neolithic Demographic Transition and its Consequences 13–34 (Springer, New York, 2008).
Gignoux, C.R., Henn, B.M. & Mountain, J.L. Rapid, global demographic expansions after the origins of agriculture. Proc. Natl. Acad. Sci. USA 108, 6044–6049 (2011).
Parwati, I., van Crevel, R. & van Soolingen, D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10, 103–111 (2010).
Barton, L. et al. Agricultural origins and the isotopic identity of domestication in northern China. Proc. Natl. Acad. Sci. USA 106, 5523–5528 (2009).
Atkinson, Q.D., Gray, R.D. & Drummond, A.J. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol. Biol. Evol. 25, 468–474 (2008).
Wei, W. et al. A calibrated human Y-chromosomal phylogeny based on resequencing. Genome Res. 23, 388–395 (2013).
Hamilton, M.J., Milne, B.T., Walker, R.S., Burger, O. & Brown, J.H. The complex structure of hunter-gatherer social networks. Proc. Biol. Sci. 274, 2195–2202 (2007).
Linz, B. et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918 (2007).
Littman, D.R. & Pamer, E.G. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10, 311–323 (2011).
Perry, S. et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS ONE 5, e8804 (2010).
de Jong, B.C. et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J. Infect. Dis. 198, 1037–1043 (2008).
Martineau, A.R. et al. Reciprocal seasonal variation in vitamin D status and tuberculosis notifications in Cape Town, South Africa. Proc. Natl. Acad. Sci. USA 108, 19013–19017 (2011).
Barnes, I., Duda, A., Pybus, O.G. & Thomas, M.G. Ancient urbanization predicts genetic resistance to tuberculosis. Evolution 65, 842–848 (2011).
Ramachandran, S. et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA 102, 15942–15947 (2005).
Quail, M.A. et al. A large genome center 's improvements to the Illumina sequencing system. Nature Methods 5, 1005–1010 (2008).
Comas, I. et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42, 498–503 (2010).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999).
Lemey, P., Rambaut, A., Drummond, A.J. & Suchard, M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5, e1000520 (2009).
Yu, Y., Harris, A.J. & He, X. S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850 (2010).
Parker, J., Rambaut, A. & Pybus, O.G. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8, 239–246 (2008).
Acknowledgements
We thank D. Behar and S. Rosset for providing the mitochondrial genome sequences and C. Gignoux for advice on the mitochondrial Neolithic data set, N. Mistry (The Foundation for Medical Research) for providing bacterial strains and C. Dye, F. Balloux and L. Weinert for comments on the manuscript. This work was supported by the MRC UK (grants U.1175.02.002.00015.01 to S.G. and U117581288 to D.Y.), the Swiss National Science Foundation (PP0033-119205 to S.G.), the US National Institutes of Health (AI090928 and HHSN266200700022C to S.G.), the Leverhulme-Royal Society Africa Award (AA080019 to S.G.) and the Natural Science Foundation of China (grant 91231115 to Q.G.). DNA sequencing was partially supported by core funding of the Wellcome Trust (grant 098051) and by a Framework Programme 7 project of the European Community (SysteMTb HEALTH-F4-2010-241587 to D.Y.). I.C. is supported by European Union funding from the Marie Curie Framework Programme 7 actions (project 272086) and project BFU2011-24112 from the Ministerio de Economía y Competitividad (Spain).
Author information
Authors and Affiliations
Contributions
I.C., Q.G., D.Y. and S.G. designed and supervised the study. M.C., S. Borrell, K.E.H., M.K.-M., J.P., B.M., S. Berg, G.T., D.Y.-M., G.B., J.M., L.W., S.R.H., S.N., R.D., A.A., Q.G. and S.G. provided MTBC strains and/or reagents. J.P., S. Bentley and S.R.H. contributed to the genome sequencing. I.C., M.C. and T.L. analyzed the data. I.C., M.C., T.L., S. Borrell, K.E.H., J.P., S. Berg, G.T., D.Y.-M., S. Bentley, S.R.H., S.N., A.A., Q.G., D.Y. and S.G. contributed to the manuscript writing. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Table 7 and Supplementary Note (PDF 2301 kb)
Supplementary Table 1
List of mycobacterial strains used in this study (XLSX 34 kb)
Supplementary Table 2
Variable single nucleotide positions in the 219 MTBC dataset (excluding outgroup) (XLSX 2760 kb)
Supplementary Table 3
Accession number and haplogroup of the 4,955 human mitochondrial (mtDNA) genomes reference dataset (XLSX 104 kb)
Supplementary Table 4
MTBC strains and human mtDNA genomes used to test for the statistical association (XLSX 13 kb)
Supplementary Table 5
Accession number, haplogroup and prehistoric period of the 423 human mtDNA genomes obtained from Gignoux et al. 2011 (XLSX 17 kb)
Supplementary Table 6
Accession number and haplogroup of the human mtDNA genomes used for the analysis of the Neolithic in East Asia (XLSX 12 kb)
Rights and permissions
About this article
Cite this article
Comas, I., Coscolla, M., Luo, T. et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45, 1176–1182 (2013). https://doi.org/10.1038/ng.2744
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2744
This article is cited by
-
Assessment of interleukin 6 (IL-6) as a marker of inflammation among adult patients with pulmonary tuberculosis in Zaria, Nigeria
The Egyptian Journal of Bronchology (2024)
-
Isolation and identification of nontuberculous mycobacteria from raw milk and traditional cheese based on the 16S rRNA and hsp65 genes, Tehran, Iran
Folia Microbiologica (2024)
-
Performance evaluation of core genome multilocus sequence typing for genotyping of Mycobacterium tuberculosis strains in China: based on multicenter, population-based collection
European Journal of Clinical Microbiology & Infectious Diseases (2024)
-
An overview of tuberculosis outbreaks reported in the years 2011–2020
BMC Infectious Diseases (2023)
-
Molecular epidemiology and drug sensitivity of Mycobacterium tuberculosis in homeless individuals in the Addis Ababa city, Ethiopia
Scientific Reports (2023)