Abstract
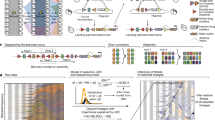
The classical model of adaptive evolution in an asexual population postulates that each adaptive clone is derived from the one preceding it1. However, experimental evidence has suggested more complex dynamics2,3,4,5, with theory predicting the fixation probability of a beneficial mutation as dependent on the mutation rate, population size and the mutation's selection coefficient6. Clonal interference has been demonstrated in viruses7 and bacteria8 but not in a eukaryote, and a detailed molecular characterization is lacking. Here we use three different fluorescent markers to visualize the dynamics of asexually evolving yeast populations. For each adaptive clone within one of our evolving populations, we identified the underlying mutations, monitored their population frequencies and used microarrays to characterize changes in the transcriptome. These results represent the most detailed molecular characterization of experimental evolution to date and provide direct experimental evidence supporting both the clonal interference and the multiple mutation models.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Muller, H.J. Some genetic aspects of sex. Am. Nat. 66, 118–138 (1932).
Helling, R.B., Vargas, C.N. & Adams, J. Evolution of Escherichia coli during growth in a constant environment. Genetics 116, 349–358 (1987).
Notley-McRobb, L. & Ferenci, T. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose limited populations of Escherichia coli. Environ. Microbiol. 1, 45–52 (1999).
Notley-McRobb, L. & Ferenci, T. Experimental analysis of molecular events during mutational periodic selections in bacterial evolution. Genetics 156, 1493–1501 (2000).
Rosenzweig, R.F., Sharp, R.R., Treves, D.S. & Adams, J. Microbial evolution in a simple unstructured environment—genetic differentiation in Escherichia coli. Genetics 137, 903–917 (1994).
Gerrish, P.J. & Lenski, R.E. The fate of competing beneficial mutations in an asexual population. Genetica 103, 127–144 (1998).
Miralles, R., Gerrish, P.J., Moya, A. & Elena, S.F. Clonal interference and the evolution of RNA viruses. Science 285, 1745–1747 (1999).
Hegreness, M., Shoresh, N., Hartl, D. & Kishony, R. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311, 1615–1617 (2006).
Crow, J.F. & Kimura, M. Evolution in sexual and asexual populations. Am. Nat. 99, 439–450 (1965).
Paquin, C. & Adams, J. Relative fitness can decrease in evolving asexual populations of S. cerevisiae. Nature 306, 368–371 (1983).
Atwood, K.C., Schneider, L.K. & Ryan, F.J. Periodic selection in Escherichia coli. Genetics 37, 146–155 (1951).
Novick, A. & Szilard, L. Experiments with the chemostat on spontaneous mutations of bacteria. Proc. Natl. Acad. Sci. USA 36, 708–719 (1950).
Paquin, C. & Adams, J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature 302, 495–500 (1983).
Kim, Y. & Orr, H.A. Adaptation in sexuals vs. asexuals: clonal interference and the Fisher-Muller model. Genetics 171, 1377–1386 (2005).
Adams, J. & Oeller, P.W. Structure of evolving populations of Saccharomyces cerevisiae adaptive changes are frequently associated with sequence alterations involving mobile elements belonging to the Ty family. Proc. Natl. Acad. Sci. USA 83, 7124–7127 (1986).
Herring, C.D. et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 38, 1406–1412 (2006).
Brown, C.J., Todd, K.M. & Rosenzweig, R.F. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 15, 931–942 (1998).
Dunham, M.J. et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99, 16144–16149 (2002).
Segre, A.V., Murray, A.W. & Leu, J.Y. High-resolution mutation mapping reveals parallel experimental evolution in yeast. PLoS Biol. 4, e256 (2006).
Blanc, V.M. & Adams, J. Evolution in Saccharomyces cerevisiae: identification of mutations increasing fitness in laboratory populations. Genetics 165, 975–983 (2003).
Gresham, D. et al. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311, 1932–1936 (2006).
Germer, S., Holland, M.J. & Higuchi, R. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res. 10, 258–266 (2000).
Wang, Y. et al. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2, e128 (2004).
de Visser, J.A.G.M. & Rozen, D.E. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172, 2093–2100 (2006).
de Visser, J.A.G.M., Zeyl, C.W., Gerrish, P.J., Blanchard, J.L. & Lenski, R.E. Diminishing returns from mutation supply rate in asexual populations. Science 283, 404–406 (1999).
Gerrish, P. The rhythm of microbial adaptation. Nature 413, 299–302 (2001).
Rozen, D.E. & de Visser, J.A.G.M. & Gerrish, P.J. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12, 1040–1045 (2002).
Desai, M.M. & Fisher, D.S. Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics 176, 1759–1798 (2007).
Desai, M.M., Fisher, D.S. & Murray, A.W. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17, 385–394 (2007).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Acknowledgements
This work was supported by US National Institutes of Health grants R01 HG003328 (G.S.) and F32 GM079113 (K.C.K.). We thank J. Ying for help with sample handling, E. Tanner for carrying out some of the gene expression microarray experiments, H. Tang for discussions on experimental design, J. Horecka for help with RT-PCR and F.Rosenzweig, B. Dunn and A. Sidow for comments on the manuscript.
Author information
Authors and Affiliations
Contributions
K.C.K. and G.S. conceived of and designed this study. K.C.K. performed all experiments and analyses. K.C.K. and G.S. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–3 and Supplementary Methods (PDF 2042 kb)
Rights and permissions
About this article
Cite this article
Kao, K., Sherlock, G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet 40, 1499–1504 (2008). https://doi.org/10.1038/ng.280
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.280
This article is cited by
-
An improved algorithm for inferring mutational parameters from bar-seq evolution experiments
BMC Genomics (2023)
-
Fit-Seq2.0: An Improved Software for High-Throughput Fitness Measurements Using Pooled Competition Assays
Journal of Molecular Evolution (2023)
-
Genetic interaction network has a very limited impact on the evolutionary trajectories in continuous culture-grown populations of yeast
BMC Ecology and Evolution (2021)
-
Evolutionary dynamics and structural consequences of de novo beneficial mutations and mutant lineages arising in a constant environment
BMC Biology (2021)
-
Changes in the distribution of fitness effects and adaptive mutational spectra following a single first step towards adaptation
Nature Communications (2021)