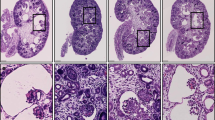
Abstract
Autosomal recessive polycystic kidney disease (ARPKD), usually considered to be a genetically homogeneous disease caused by mutations in PKHD1, has been associated with ciliary dysfunction. Here, we describe mutations in DZIP1L, which encodes DAZ interacting protein 1-like, in patients with ARPKD. We further validated these findings through loss-of-function studies in mice and zebrafish. DZIP1L localizes to centrioles and to the distal ends of basal bodies, and interacts with septin2, a protein implicated in maintenance of the periciliary diffusion barrier at the ciliary transition zone. In agreement with a defect in the diffusion barrier, we found that the ciliary-membrane translocation of the PKD proteins polycystin-1 and polycystin-2 is compromised in DZIP1L-mutant cells. Together, these data provide what is, to our knowledge, the first conclusive evidence that ARPKD is not a homogeneous disorder and further establish DZIP1L as a second gene involved in ARPKD pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Guay-Woodford, L.M. et al. Consensus expert recommendations for the diagnosis and management of autosomal recessive polycystic kidney disease: report of an international conference. J. Pediatr. 165, 611–617 (2014).
Bergmann, C. et al. Clinical consequences of PKHD1 mutations in 164 patients with autosomal-recessive polycystic kidney disease (ARPKD). Kidney Int. 67, 829–848 (2005).
Habbig, S. & Liebau, M.C. Ciliopathies: from rare inherited cystic kidney diseases to basic cellular function. Mol Cell Pediatr 2, 8 (2015).
Garcia-Gonzalo, F.R. et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776–784 (2011).
Jensen, V.L. et al. Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J. 34, 2537–2556 (2015).
Reiter, J.F., Blacque, O.E. & Leroux, M.R. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13, 608–618 (2012).
Guay-Woodford, L.M. Autosomal recessive polycystic kidney disease: the prototype of the hepato-renal fibrocystic diseases. J. Pediatr. Genet. 3, 89–101 (2014).
Wang, S., Luo, Y., Wilson, P.D., Witman, G.B. & Zhou, J. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J. Am. Soc. Nephrol. 15, 592–602 (2004).
Zhang, M.Z. et al. PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc. Natl. Acad. Sci. USA 101, 2311–2316 (2004).
Wu, Y. et al. Kinesin-2 mediates physical and functional interactions between polycystin-2 and fibrocystin. Hum. Mol. Genet. 15, 3280–3292 (2006).
Wang, S. et al. Fibrocystin/polyductin, found in the same protein complex with polycystin-2, regulates calcium responses in kidney epithelia. Mol. Cell. Biol. 27, 3241–3252 (2007).
Kim, I. et al. Fibrocystin/polyductin modulates renal tubular formation by regulating polycystin-2 expression and function. J. Am. Soc. Nephrol. 19, 455–468 (2008).
Delling, M., DeCaen, P.G., Doerner, J.F., Febvay, S. & Clapham, D.E. Primary cilia are specialized calcium signalling organelles. Nature 504, 311–314 (2013).
DeCaen, P.G., Delling, M., Vien, T.N. & Clapham, D.E. Direct recording and molecular identification of the calcium channel of primary cilia. Nature 504, 315–318 (2013).
Delling, M. et al. Primary cilia are not calcium-responsive mechanosensors. Nature 531, 656–660 (2016).
Eisenberger, T. et al. An efficient and comprehensive strategy for genetic diagnostics of polycystic kidney disease. PLoS One 10, e0116680 (2015).
Glazer, A.M. et al. The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev. Biol. 337, 148–156 (2010).
Tay, S.Y. et al. The iguana/DZIP1 protein is a novel component of the ciliogenic pathway essential for axonemal biogenesis. Dev. Dyn. 239, 527–534 (2010).
Wolff, C. et al. iguana encodes a novel zinc-finger protein with coiled-coil domains essential for Hedgehog signal transduction in the zebrafish embryo. Genes Dev. 18, 1565–1576 (2004).
Sekimizu, K. et al. The zebrafish iguana locus encodes Dzip1, a novel zinc-finger protein required for proper regulation of Hedgehog signaling. Development 131, 2521–2532 (2004).
Kim, H.R., Richardson, J., van Eeden, F. & Ingham, P.W. Gli2a protein localization reveals a role for Iguana/DZIP1 in primary ciliogenesis and a dependence of Hedgehog signal transduction on primary cilia in the zebrafish. BMC Biol. 8, 65 (2010).
Wang, C., Low, W.C., Liu, A. & Wang, B. Centrosomal protein DZIP1 regulates Hedgehog signaling by promoting cytoplasmic retention of transcription factor GLI3 and affecting ciliogenesis. J. Biol. Chem. 288, 29518–29529 (2013).
Papathanasiou, P. et al. A recessive screen for genes regulating hematopoietic stem cells. Blood 116, 5849–5858 (2010).
Huangfu, D. & Anderson, K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA 102, 11325–11330 (2005).
Litingtung, Y., Dahn, R.D., Li, Y., Fallon, J.F. & Chiang, C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418, 979–983 (2002).
Perner, B., Englert, C. & Bollig, F. The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 309, 87–96 (2007).
Kobayashi, T. & Dynlacht, B.D. Regulating the transition from centriole to basal body. J. Cell Biol. 193, 435–444 (2011).
Hu, Q. et al. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329, 436–439 (2010).
Chih, B. et al. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14, 61–72 (2011).
Ong, A.C. et al. Polycystin-1 expression in PKD1, early-onset PKD1, and TSC2/PKD1 cystic tissue. Kidney Int. 56, 1324–1333 (1999).
Bergmann, C. et al. Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1). J. Am. Soc. Nephrol. 14, 76–89 (2003).
Bergmann, C. et al. PKHD1 mutations in autosomal recessive polycystic kidney disease (ARPKD). Hum. Mutat. 23, 453–463 (2004).
Gunay-Aygun, M. et al. Correlation of kidney function, volume and imaging findings, and PKHD1 mutations in 73 patients with autosomal recessive polycystic kidney disease. Clin. J. Am. Soc. Nephrol. 5, 972–984 (2010).
Nakanishi, K., Sweeney, W.E. Jr., Zerres, K., Guay-Woodford, L.M. & Avner, E.D. Proximal tubular cysts in fetal human autosomal recessive polycystic kidney disease. J. Am. Soc. Nephrol. 11, 760–763 (2000).
Williams, S.S., Cobo-Stark, P., James, L.R., Somlo, S. & Igarashi, P. Kidney cysts, pancreatic cysts, and biliary disease in a mouse model of autosomal recessive polycystic kidney disease. Pediatr. Nephrol. 23, 733–741 (2008).
Bakeberg, J.L. et al. Epitope-tagged Pkhd1 tracks the processing, secretion, and localization of fibrocystin. J. Am. Soc. Nephrol. 22, 2266–2277 (2011).
Woollard, J.R. et al. A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int. 72, 328–336 (2007).
Davis, E.E. & Katsanis, N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr. Opin. Genet. Dev. 22, 290–303 (2012).
Boulter, C. et al. Cardiovascular, skeletal, and renal defects in mice with a targeted disruption of the Pkd1 gene. Proc. Natl. Acad. Sci. USA 98, 12174–12179 (2001).
Kolpakova-Hart, E. et al. Growth of cranial synchondroses and sutures requires polycystin-1. Dev. Biol. 321, 407–419 (2008).
Rossi, A. et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233 (2015).
Blum, M., De Robertis, E.M., Wallingford, J.B. & Niehrs, C. Morpholinos: antisense and Sensibility. Dev. Cell 35, 145–149 (2015).
Zhang, B. et al. GSK3β-Dzip1-Rab8 cascade regulates ciliogenesis after mitosis. PLoS Biol. 13, e1002129 (2015).
Diener, D.R., Lupetti, P. & Rosenbaum, J.L. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr. Biol. 25, 379–384 (2015).
Toriyama, M. et al. The ciliopathy-associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat. Genet. 48, 648–656 (2016).
Ma, M., Tian, X., Igarashi, P., Pazour, G.J. & Somlo, S. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat. Genet. 45, 1004–1012 (2013).
Braun, D.A. et al. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat. Genet. 48, 457–465 (2016).
Zhou, W. et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat. Genet. 44, 910–915 (2012).
Hoff, S. et al. ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat. Genet. 45, 951–956 (2013).
Andrews, T.D. et al. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol. 2, 120061 (2012).
Rohatgi, R., Milenkovic, L. & Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–376 (2007).
Bruce, S.J. et al. Inactivation of Patched1 in the mouse limb has novel inhibitory effects on the chondrogenic program. J. Biol. Chem. 285, 27967–27981 (2010).
Fowles, L.F. et al. Genomic screen for genes involved in mammalian craniofacial development. Genesis 35, 73–87 (2003).
Butterfield, N.C. et al. Patched 1 is a crucial determinant of asymmetry and digit number in the vertebrate limb. Development 136, 3515–3524 (2009).
Ashe, A. et al. Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Hum. Mol. Genet. 21, 1808–1823 (2012).
Todaro, G.J. & Green, H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17, 299–313 (1963).
Shibazaki, S. et al. Cyst formation and activation of the extracellular regulated kinase pathway after kidney specific inactivation of Pkd1. Hum. Mol. Genet. 17, 1505–1516 (2008).
Grimm, D.H. et al. Polycystin-1 distribution is modulated by polycystin-2 expression in mammalian cells. J. Biol. Chem. 278, 36786–36793 (2003).
Wu, G. et al. Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell 93, 177–188 (1998).
Xie, W., Horn, H.F. & Wright, G.D. Superresolution microscopy of the nuclear envelope and associated proteins. Methods Mol. Biol. 1411, 83–97 (2016).
Metzis, V. et al. Patched1 is required in neural crest cells for the prevention of orofacial clefts. Hum. Mol. Genet. 22, 5026–5035 (2013).
Kimmel, C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B. & Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Montague, T.G., Cruz, J.M., Gagnon, J.A., Church, G.M. & Valen, E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 (2014).
Bassett, A.R., Tibbit, C., Ponting, C.P. & Liu, J.L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228 (2013).
Yu, C., Zhang, Y., Yao, S. & Wei, Y. A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS One 9, e98282 (2014).
Acknowledgements
The authors thank the patients and their families for their cooperation and interest in the study; M.T. Toh and Y.T. Koh for technical assistance; W.I. Goh of the Institute of Medical Biology's Microscopy Unit for assistance with super-resolution microscopy; J. Lefevre and N. Hamilton for advice on statistical analyses; and C. Cortés, M. Pitt, F. Olsson, L. Zhao, L. Wilkinson and P. Karaith Oliva for assistance, insightful discussion and advice. The authors also thank C. Has (University Medical Center Freiburg) for kindly providing control fibroblasts; S. Somlo, M. Ma and K. Dong (Yale University) for Pkd1- and Pkd2-mutant cells; L. Lei (Nanyang Technological University) for the RPE-1 cell line stably expressing Arl13b–GFP; L. Pelletier (Lunenfeld-Tanenbaum Research Institute) for basal-body and transition-zone-protein cDNA clones; and R. Witzgall (Institute for Molecular and Cellular Anatomy, University of Regensburg), G. Wu (Center of Translational Cancer Research and Therapy, Beijing) and R. Rohatgi (Stanford University) for antibodies. We also thank the staff of the University of Queensland (UQ) QBP animal house for assistance with mouse husbandry and the Australian Phenomics Facility for maintaining mice throughout the screen. Confocal microscopy at UQ was carried out at the Institute for Molecular Bioscience Dynamic Imaging Facility for Cancer Biology, which is funded through the generous support of the Australian Cancer Research Foundation. We also acknowledge the Australian Microscopy & Microanalysis Facility (AMMRF) at the Centre for Microscopy and Microanalysis at UQ. M.H., S.N., V.F. and C.B. are supported as employees of Bioscientia/Sonic Healthcare; C.W. was supported as a recipient of a University of Queensland Vice-Chancellor's Senior Research Fellowship; and S.R. is supported as a Senior Principal Investigator at the Institute of Molecular and Cell Biology, Singapore. M.H.L. is supported as a Senior Principal Research Fellow of the Australian National Health and Medical Research Council (NHMRC). F.H. is supported as the Warren E. Grupe Professor. This work was supported by grants from the German Research Fund (DFG) to K.Z. and C.B., the DFG Collaborative Research Centre (SFB) KIDGEM 1140 and the Federal Ministry of Education and Research (BMBF, 01GM1515C) to C.B., the Australian NHMRC (APP1045464) to C.W., the National Institutes of Health NIH (DK068306) to F.H. and the Agency for Science, Technology and Research (A*STAR) of Singapore to W.H. and S.R. This paper is dedicated to the memory of Markus Nauck, who recently passed away.
Author information
Authors and Affiliations
Contributions
H.L., P.J.K., S.Y.T., S.V. and S.R. performed the zebrafish mutant and morphant analyses, protein interaction studies, localization experiments with human fibroblasts and validation of antibodies to PCs. W.H. supervised the protein interaction studies. E.O., C.K. and D.E. performed the zebrafish morpholino analyses. M.C.R.G., G.K., V.M. and A.D.C. performed the mouse analyses, and M.C.R.G. and G.K. performed the localization experiments with mouse cells. R.T., P.P. and A.C.P. performed the screen that identified the mouse mutant, and B.W. was involved in mapping and identifying the mouse mutation. G.D.W. performed the super-resolution microscopy experiments. M.H.L. assisted with design and analysis of mouse kidney experiments. N.O.-B., N.H., V.F. and S.N. performed the human mutation analysis. S.N., V.F., M.H., H.Y.G., E.A.O., F.H. and C.B. carried out the WES data processing and analyses. S.N., V.F., E.W., U.V., H.Y.G., K.Z., F.H. and C.B. recruited and clinically characterized the study subjects and collected samples. B.H. performed histologic evaluation of the data. S.R., C.W. and C.B. conceived the project, designed and supervised the experiments, analyzed and interpreted the data and wrote the manuscript. All authors reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Pedigree and DZIP1L mutation data.
Top: Pedigrees and sequence electropherograms of the patients described in this study depicting the respective homozygous DZIP1L mutations. Bottom: Renal ultrasound data of patients with DZIP1L mutations demonstrating bilaterally enlarged, hyperechogenic kidneys with poor cortico-medullary differentiation and multiple tiny cysts characteristic of ARPKD. (a-e) Longitudinal sections of right (a, patient B16-3; d, patient A3533-1; e, patient B8031-1) and left (b,c, patient B16-2) kidney with a large number of echogenic spots and tiny, predominantly medullary cysts throughout the kidney (“pepper-salt pattern”). (f,g) Transverse (f) and longitudinal (g) section of the left kidney of patient B8031-1 with multiple small cysts and echogenic spots. Color doppler with multiple twinkling phenomena identifying these spots as small calcifications (g).
Supplementary Figure 2 Sequence alignment of human, mouse and zebrafish DZIP1L proteins.
Multiple sequence alignment of human (Accession number: NM_173543.2), mouse (Accession number: NM_028258.4) and zebrafish DZIP1L proteins (Accession number: NM_001037227.1) using Clustal W method. Amino acid identity is shaded (black). The zinc finger (red) and coiled-coil domains (blue) are highlighted.
Supplementary Figure 3 Characterization of Dzip1lwpy/wpy mice.
(a) Sanger sequencing scans of the wild-type (top) and homozygous Dzip1lwpy/wpy mutation (bottom), and the location of the stop codon in the predicted protein (on the right). Asterisks mark the mutated base, arrow marks location of the stop codon. (b) Dzip1lwpy/wpy mutant embryos show evidence of altered hedgehog signalling in the limb. Indicated markers analysed by wholemount in situ hybridisation in 11.5 dpc limbs show expanded ectopic expression of Hoxd13, Grem1 and Gli1 into the anterior of the Dzip1lwpy/wpy mutant forelimb bud (arrows in b). Anterior is to the top in all limb images. Similar data were obtained in hindlimbs (not shown). n = limbs from 3 independent embryos. (c) IF analysis with a battery of markers to hedgehog-dependent neural populations reveals no consistent difference between wild-type control and Dzip1lwpy/wpy mutant sections through the neural tube of 10.5 dpc embryos at the level of the forelimb. Similar data were obtained with sections at hindlimb level (not shown). n = multiple sections from 3 independent embryos for all markers except Shh (n=2). Scale bar in b = 200μm; c = 100μm. (d) qRT-PCR analysis of Gli1 transcript levels shows that, compared to Dzip1l+/+ MEFs, Dzip1lwpy/wpy MEFs fail to significantly upregulate HH signalling in response to SAG treatment (n=4 cell lines derived from independent embryos, cells treated in three separate experiments; each data point is the average of 3 separate culture wells). Statistical analysis based on separate unpaired two-tailed Student’s t-tests for Dzip1l+/+ t(6)=14.91, ****p<0.0001, and Dzip1lwpy/wpy t(6)=0.6943, p=0.5135, ns, not significant, +/-SAG treatment. Error bars, s.e.m. All data presented are from embryos on a C57BL/6:C3H mixed background.
Supplementary Figure 4 Examples of histological analyses of liver sections.
(a-f) Elastica van Gieson (EvG) and (g-l) Silver staining of liver sections from Dzip1l+/+ (a-c; g-j) and Dzip1lwpy/wpy (d-f; j-l) mice at P21, following at least 4 crosses to a CD1 background. Arrows mark bile ducts, showing an excess of bile ducts surrounding blood vessels in Dzip1lwpy/wpy mutant mice. Scale bar = 50μm.
Supplementary Figure 5 Maternal expression of dzip1l, efficacy of the dzip1l-splice morpholinos and generation of the dzip1l-deletion allele.
(a) The maternal expression of dzip1l mRNA at the 64-cell stage of zebrafish embryonic development. Zygotic expression is shown for the 10 somite stage. acta1b expression was used as loading control. (b) Splice morpholino (e5i5MO) efficiently interferes with dzip1l pre-mRNA splicing. RT-PCR with primers binding to exon2 and exon10, expected band of about 650 bp is visible from cDNA of wild-type embryos (arrow). A smaller band of 350 bp (arrowhead) is visible from cDNA of the morphants. Sequencing of this band showed deletion of exon 5-7. The lower band (asterisk) is a non-specific band. acta1b expression was used as loading control. (c), Splice morpholino (i1e2MO) interference with dzip1l pre-mRNA splicing. RT-PCR with primers binding to exon1 and exon3, expected band of about 680 bp is visible from cDNA of wild-type embryos (arrow) and significantly reduced in cDNA of morphant embryos. Wild-type control band was confirmed by sequencing, lower bands are unspecific. (d) Splice morpholino (i4e5MO) interferes with dzip1l pre-mRNA splicing. RT-PCR with primers binding to exon2 and exon10, expected 870 bp band is visible in cDNA of wild-type embryos and missing in morphant embryos (arrow). eef1a1l1 expression was used as loading control. (e) Schematic diagram of CRISPR/Cas9 mediated genomic modification of the dzip1l locus. Two guide RNAs, targeting exon2 and exon13, were designed to excise the intervening genomic DNA, to create a deletion allele. (f) PCR screening of CRISPR/Cas9 mediated dzip1l deletion among F0 founder fish with primers binding to exon2 and exon14. Fish 1, 2 and 3 did not carry the dzip1l deletion allele. Fish 4 and 5 were identified as dzip1l deletion allele carriers with the expected PCR product of 350 bp (arrow). Sequencing of this band showed the expected deletion. (g) Three otoliths in the otic vesicle of a 24 hpf maternal-zygotic dzip1l mutant embryo (n = 10 embryos).
Supplementary Figure 6 dzip1l zebrafish morphants display ciliopathy-associated phenotypes.
(a) Wholemount of a 20 hpf zebrafish embryo showing dzip1l expression in the nasal placode (short arrow) and pronephric duct (long arrow). (b) dzip1l expression in MCC precursors in the pronephric duct at 24 hpf (arrows). Inset shows expression in individual MCC precursors. (c) dzip1l expression in the nasal placode at 24 hpf (arrows). (d) Wild-type control embryo at 3 dpf. (e) Two otoliths in the otic vesicle of a wild-type embryo at 3 dpf. (f) dzip1l translation blocking morpholino (dzip1l MO) injected embryo at 3 dpf with shortened, slightly curved body, hydrocephalus (arrow) and reduced retinal size (arrowhead) (g) Three otoliths in the otic vesicle of a dzip1l morpholino (i4e5) injected embryo at 3 dpf. (h) Wild-type glomeruli (upper left panel) and cystic dilated glomeruli in dzip1l MO injected embryos (lower left panel, white asterisk) at 48 hpf. (i) Incidence of glomerular cysts and otolith deposition defects in control (co MO), translation blocking (TB MO) and splice site (i1e2 MO and i4e5 MO) morpholino injected Tg(wt1b::GFP) embryos at 48 hpf (for glomerular analysis n = 425 embryos for translation blocking morpholino, n = 349 embryos (i1e2) and n = 384 embryos (i4e5) for splice morpholinos; t(13)=5.68, ***p=0.0008 (i1e2 MO), t(17)=8.36 and t(14)=8.27, ****p <0.0001 (TB and i4e5 MO); for otolith analysis n = 243 embryos for translation blocking morpholino, n = 236 embryos (i1e2) and n = 326 embryos (i4e5) for splice morpholinos; t(7)=5.42, **p=0.0011 (TB MO), t(6)=14.93, ****p<0.0001(i1e2 MO), t(7)=4.34, **p=0.003 (i4e5MO). Statistical analysis based on an unpaired two-tailed Student`s t-test. Error bars, s.e.m. (j) Co-injection of zebrafish dzip1l mRNA (ddzip1l) can rescue dzip1l morpholino caused phenotypes; n = 463 embryos for morpholino and n = 313 embryos for mRNA co-injected embryos; t(9)=9.06, ****p<0.0001. Scale bars in a-g = 100μm; h = 50μm; each data point represents a single experiment.
Supplementary Figure 7 Analysis of protein and transcript levels in DZIP1L-mutant cells.
a,b) Staining with a C-terminal DZIP1L antibody (Sigma; green) reveals no signal in Dzip1lwpy/wpy MEFs (arrows in b), consistent with the truncated nature of the protein; (c,d,) Staining with an antibody raised to the entire DZIP1L protein (Abnova; green) reveals a residual level of truncated protein in Dzip1lwpy/wpy MEFs (arrows in d). The cilium is marked with acetylated-α-tubulin and γ-tubulin in (a,b; magenta) and IFT88 in (c,d; magenta). (e) Based on an unpaired two-tailed Student’s t-test, qRT-PCR revealed significantly reduced but not absent Dzip1l transcript levels in Dzip1lwpy/wpy MEFs relative to Dzip1l+/+ wild-type MEFs, t(6)=5.905, **p=0.001 (n=4, consisting of 3 cell lines derived from independent embryos, analysed across two experiments (separate RNA preparations from one cell line analysed in both experiments); each data point is the average of 3 separate culture wells). Dzip1 levels were not altered, t(6)=0.385, p=0.7135, ns, not significant, to compensate for reduced Dzip1l. Error bars, s.e.m. (f) DZIP1L (magenta) localizes at the transition zone in control human dermal fibroblasts, but is not detectable in DZIP1L mutant dermal fibroblasts from individual B155 (p.Gln155*) with the C-terminal Sigma antibody (arrows in g). Ciliary axonemes and basal bodies were labelled with anti-acetylated tubulin and γ-tubulin antibodies, respectively (green). (h) Immunoblotting with anti-DZIP1L antibody (Abnova) revealed a band corresponding to full length DZIP1L in control cell lysate (arrow) but not in lysate from DZIP1L mutant human fibroblasts. Scale bars in a-d = 2.5μm; g,h = 1μm.
Supplementary Figure 8 Dzip1lwpy/wpy-mutant embryos show defects in ciliogenesis ex vivo but not in vivo.
(a) Staining for cilia in serum-starved MEFs revealed a defect in the ability of isolated Dzip1lwpy/wpy mutant cells to ciliate relative to wild-type cells. Image and quantification based on ARL13B marking the cilium (red, arrows in a), but a significant reduction was also seen with acetylated-α-tubulin staining (n = 6 cell lines, each from an independent embryo; between 100-540 cells counted per cell line (1434 Dzip1l+/+ and 1837 Dzip1lwpy/wpy cells in total), across two separate experiments). (b) Imaging and quantification of cilia in 11.5dpc forelimb buds following staining with acetylated-α-tubulin revealed no significant difference in cilia frequency (n = forelimb from 3 independent embryos, with 9 sections analysed per limb, >1900 cells counted per limb). Note the final cilia/nuclei ratio in limb sections will not necessarily represent the actual percentage of ciliated cells in the tissue, but is a relative value across the samples (see online methods for details). Statistical analysis based on an unpaired, two-tailed Student’s t-test. (a) t(10)=10.87, ****p<0.0001; (b) t(4)=0.63, p=0.5624, ns, not significant. Error bars, s.e.m. Scale bar in a,b = 10μm. All data presented are from embryos on a C57BL/6:C3H mixed background.
Supplementary Figure 9 DZIP1L specifically associates with the ciliary-transition-zone protein SEPT2.
(a) Co-IP experiments from HEK293T cells transfected with expression constructs for epitope-tagged SEPT2 (C-terminal HA) and DZIP1L, CEP89, B9D1, MKS1 and NPHP1 (N-terminal Myc), reveal a specific interaction between SEPT2 and DZIP1L but none of the other proteins analysed. (b) Co-IP with DZIP1L (C-terminal HA) and SEPT2, CEP89, B9D1, MKS1 and NPHP1 (N-terminal Myc) confirmed the DZIP1L-SEPT2 interaction, and showed that none of the other proteins analysed interact with DZIP1L. Co-IP experiments were repeated three times.
Supplementary Figure 10 Validation of the 7e12 antibody to PC1.
(a,b) PC1 antibody (mAb 7e12 raised to the N-terminal leucine rich repeat region of PC1 (24-180aa), aliquots obtained from both Abcam and Santa Cruz Biotechnology – see online methods for details) detected overexpressed PC1 protein (green) in IMCD3 cells. (c-j) PC1 staining (green) on the ciliary membrane in Pkd1 or Pkd2 heterozygote control cells, and lack of staining in null mutant cells, with the PC1 7e12 antibody preparation indicated. Acetylated-α-tubulin (magenta) marks the ciliary axoneme. (k,l) Quantification of the percentage of ciliated cells with PC1 along the axoneme in Pkd1 or Pkd2 heterozygous control cells. No homozygous mutant cells analysed displayed detectable PC1 staining along the axoneme. Note that PC2 is required for ciliary trafficking of PC1, so axonemal PC1 staining would not be expected in Pkd2 null cells. Cilia were scored from two independent experiments. Approximately 100 ciliated cells were analysed in each experiment. The values are not compared statistically. Error bars, s.e.m. Scale bars = 1μm. See online methods for details of the Pkd1 and Pkd2 cell lines used for these validation experiments.
Supplementary Figure 11 Validation of antibody to PC2 and localization of SMO in Dzip1lwpy/wpy MEFs.
(a, b) PC2 staining (green) on the ciliary membrane in Pkd2 heterozygote control but not null mutant cells with the PC2 antibody (MV12). Acetylated-α-tubulin (magenta) marks the ciliary axoneme. (c) Quantification of the percentage of ciliated cells with PC2 along the axoneme in Pkd2 heterozygote and homozygote mutant cells. Statistical analysis based on an unpaired two-tailed Student’s t-test, t(5)=2.57,**p=0.0033. Cilia were scored in three experiments, with cells from two independent coverslips analysed for each experiment (approximately 100 ciliated cells scored on each coverslip). (d,e) In response to treatment with the HH signalling agonist SAG, SMO (green) localizes along the axoneme (acetylated-α-tubulin, magenta) in both Dzip1l+/+ and Dzip1lwpy/wpy MEFs. (f) quantification shows no difference in SMO localization between the two genotypes (n=4 MEF cell lines derived from individual embryos, analysed across two experiments; 28-91 cilia scored per cell line, total 299 Dzip1l+/+ cilia and 137 Dzip1lwpy/wpy cilia scored). Statistical analysis based on an unpaired two-tailed Student’s t-test, t(6)=0.0292, p=0.9776, ns, not significant. Error bars, s.e.m. Scale bars in a,b = 1μm; d,e = 2μm.
Supplementary Figure 12 Comparison of REVEL and CADD pathogenicity-prediction scores between DZIP1L and PKHD1.
A higher score indicates a more likely pathogenic effect of nucleotide exchanges at the corresponding position. For DZIP1L, red bars denote SNVs reported in this study, and green bars denote missense or loss-of-function variants found in ExAC. Height of green bars corresponds to the ExAC allele frequency in percent. (a) REVEL score. The x-axis refers to the coding DNA position of the longest transcript of the respective gene (DZIP1L = ENST00000327532; PKHD1 = ENST00000371117). Exon numbers and bounds are drawn above the axis. The y-axis displays scores for all possible amino acid changes at the corresponding position, averaged with a scope of 4 bases to the left and right by loess smoothing (b) CADD score. The x-axis refers to the coding DNA position of the longest transcript of the respective gene (DZIP1L = ENST00000327532; PKHD1 = ENST00000371117). Exon numbers and bounds are drawn above the axis. The y-axis displays scores for all possible amino acid changes at the corresponding position, averaged with a scope of 3 bases to the left and right by application of a Daniell kernel function.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 and Supplementary Tables 1 and 3. (PDF 2700 kb)
Supplementary Table 2
DZIP1L Y2H screen. (XLS 26 kb)
Rights and permissions
About this article
Cite this article
Lu, H., Galeano, M., Ott, E. et al. Mutations in DZIP1L, which encodes a ciliary-transition-zone protein, cause autosomal recessive polycystic kidney disease. Nat Genet 49, 1025–1034 (2017). https://doi.org/10.1038/ng.3871
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3871
This article is cited by
-
Transport and barrier mechanisms that regulate ciliary compartmentalization and ciliopathies
Nature Reviews Nephrology (2024)
-
Emerging mechanistic understanding of cilia function in cellular signalling
Nature Reviews Molecular Cell Biology (2024)
-
Epidemiology and outcomes of pediatric autosomal recessive polycystic kidney disease in the Middle East and North Africa
Pediatric Nephrology (2024)
-
Identification of genetic variants associated with anterior cruciate ligament rupture and AKC standard coat color in the Labrador Retriever
BMC Genomic Data (2023)
-
Design of two ongoing clinical trials of tolvaptan in the treatment of pediatric patients with autosomal recessive polycystic kidney disease
BMC Nephrology (2023)