Abstract
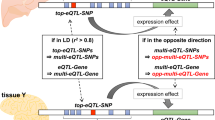
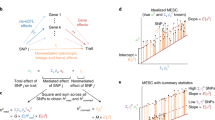
How to interpret the biological causes underlying the predisposing markers identified through genome-wide association studies (GWAS) remains an open question. One direct and powerful way to assess the genetic causality behind GWAS is through analysis of expression quantitative trait loci (eQTLs). Here we describe a new approach to estimate the tissues behind the genetic causality of a variety of GWAS traits, using the cis-eQTLs in 44 tissues from the Genotype-Tissue Expression (GTEx) Consortium. We have adapted the regulatory trait concordance (RTC) score to measure the probability of eQTLs being active in multiple tissues and to calculate the probability that a GWAS-associated variant and an eQTL tag the same functional effect. By normalizing the GWAS–eQTL probabilities by the tissue-sharing estimates for eQTLs, we generate relative tissue-causality profiles for GWAS traits. Our approach not only implicates the gene likely mediating individual GWAS signals, but also highlights tissues where the genetic causality for an individual trait is likely manifested.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bush, W.S. & Moore, J.H. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 8, e1002822 (2012).
McCarthy, M.I. et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369 (2008).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Dermitzakis, E.T. From gene expression to disease risk. Nat. Genet. 40, 492–493 (2008).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Nica, A.C. & Dermitzakis, E.T. Using gene expression to investigate the genetic basis of complex disorders. Hum. Mol. Genet. 17 R2, R129–R134 (2008).
Montgomery, S.B. & Dermitzakis, E.T. From expression QTLs to personalized transcriptomics. Nat. Rev. Genet. 12, 277–282 (2011).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Flutre, T., Wen, X., Pritchard, J. & Stephens, M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 9, e1003486 (2013).
Brown, C.D., Mangravite, L.M. & Engelhardt, B.E. Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet. 9, e1003649 (2013).
Hore, V. et al. Tensor decomposition for multiple-tissue gene expression experiments. Nat. Genet. 48, 1094–1100 (2016).
Nica, A.C. et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 6, e1000895 (2010).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Dimas, A.S. et al. Common regulatory variation impacts gene expression in a cell type–dependent manner. Science 325, 1246–1250 (2009).
Gutierrez-Arcelus, M. et al. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 11, e1004958 (2015).
Nicolae, D.L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888 (2010).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Nguyen, P. et al. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. (Berl.) 92, 272–283 (2008).
Saltiel, A.R. & Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 (2001).
Maiuolo, J., Oppedisano, F., Gratteri, S., Muscoli, C. & Mollace, V. Regulation of uric acid metabolism and excretion. Int. J. Cardiol. 213, 8–14 (2016).
Taki, Y. et al. Correlation among body height, intelligence, and brain gray matter volume in healthy children. Neuroimage 59, 1023–1027 (2012).
Buckner, R.L., Andrews-Hanna, J.R. & Schacter, D.L. The brain's default network: anatomy, function, and relevance to disease. Ann. NY Acad. Sci. 1124, 1–38 (2008).
Harrison, P.J. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain 122, 593–624 (1999).
Han, S.K. et al. Activation of gonadotropin-releasing hormone neurons by kisspeptin as a neuroendocrine switch for the onset of puberty. J. Neurosci. 25, 11349–11356 (2005).
Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 (2009).
Samani, N.J. et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 357, 443–453 (2007).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Musunuru, K. et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–719 (2010).
Suzuki, R. & Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006).
GTEx Consortium. Genetic effects on gene expression across human tissues. Nature http://dx.doi.org/10.1038/nature24277 (2017).
Ongen, H., Buil, A., Brown, A.A., Dermitzakis, E.T. & Delaneau, O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485 (2016).
Delaneau, O. et al. A complete tool set for molecular QTL discovery and analysis. Nat. Commun. 8, 15452 (2017).
McVean, G.A. et al. The fine-scale structure of recombination rate variation in the human genome. Science 304, 581–584 (2004).
Kent, W.J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Su, Z., Marchini, J. & Donnelly, P. HAPGEN2: simulation of multiple disease SNPs. Bioinformatics 27, 2304–2305 (2011).
Acknowledgements
This research was supported by grants from the US National Institutes of Health (NIH-R01MH101814), European Commission Framework Programme 7 (UE7-SYSCOL-258236), the European Research Council (UE7-POPRNASEQ-260927), the Swiss National Science Foundation (31003A-149984 and 31003A-170096), and the Louis Jeantet Foundation. Computations were performed at the Vital-IT Centre for High-Performance Computing of the SIB Swiss Institute of Bioinformatics.
Author information
Authors and Affiliations
Consortia
Contributions
H.O. and E.T.D. designed the study. H.O., A.A.B., and O.D. conducted the analysis and developed software. A.C.N. designed the original RTC method. N.I.P. tested the software. H.O. wrote and E.T.D. edited the manuscript. The GTEx Consortium generated the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members and affiliations appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–19 and Supplementary Note (PDF 1417 kb)
Supplementary Table 1
Mean eQTL tissue sharing probabilities across 44 tissues. (XLSX 35 kb)
Supplementary Table 2
Proportions of the number of tissues an eQTL (FDR = 5%) is active in, for the 44 GTEx tissues. (XLSX 31 kb)
Supplementary Table 3
Frequency of tissues being included in the most likely set of tissues for all eQTLs discovered. (XLSX 33 kb)
Supplementary Table 4
Various error statistics for r2. (XLSX 17 kb)
Supplementary Table 5
Enrichment over the null for the GWAS traits. (XLSX 315 kb)
Supplementary Table 6
Nominal P values for enrichments over the null for the GWAS traits. (XLSX 261 kb)
Supplementary Table 7
Normalized tissue causality profile for the GWAS traits. (XLSX 333 kb)
Supplementary Table 8
GWAS–eQTL probabilities. (XLSX 1923 kb)
Rights and permissions
About this article
Cite this article
Ongen, H., Brown, A., Delaneau, O. et al. Estimating the causal tissues for complex traits and diseases. Nat Genet 49, 1676–1683 (2017). https://doi.org/10.1038/ng.3981
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3981
This article is cited by
-
Non-coding autoimmune risk variant defines role for ICOS in T peripheral helper cell development
Nature Communications (2024)
-
A landscape of gene expression regulation for synovium in arthritis
Nature Communications (2024)
-
Examination of a novel expression-based gene-SNP annotation strategy to identify tissue-specific contributions to heritability in multiple traits
European Journal of Human Genetics (2024)
-
An overview of detecting gene-trait associations by integrating GWAS summary statistics and eQTLs
Science China Life Sciences (2024)
-
Analysis of genetically determined gene expression suggests role of inflammatory processes in exfoliation syndrome
BMC Genomics (2023)