Abstract
Focal segmental glomerulosclerosis (FSGS) is a pattern of kidney injury observed either as an idiopathic finding or as a consequence of underlying systemic conditions. Several genes have been identified that, when mutated, lead to inherited FSGS and/or the related nephrotic syndrome. These findings have accelerated the understanding of glomerular podocyte function and disease, motivating our search for additional FSGS genes. Using linkage analysis, we identified a locus for autosomal-dominant FSGS susceptibility on a region of chromosome 14q. By sequencing multiple genes in this region, we detected nine independent nonconservative missense mutations in INF2, which encodes a member of the formin family of actin-regulating proteins. These mutations, all within the diaphanous inhibitory domain of INF2, segregate with FSGS in 11 unrelated families and alter highly conserved amino acid residues. The observation that alterations in this podocyte-expressed formin cause FSGS emphasizes the importance of fine regulation of actin polymerization in podocyte function.
Similar content being viewed by others
Main
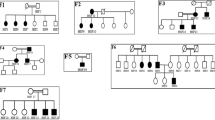
We genotyped members of a large family with apparent autosomal dominant FSGS using a dense set of SNP markers (Affymetrix 250K SNP Chip array) followed by focused microsatellite genotyping. We identified a locus for FSGS in this family on 14q32 (family FGJN; Fig. 1 and Supplementary Fig. 1). This region overlapped with a larger locus that we previously identified through linkage analysis in another large family with a similar clinical course (family FGBR; Fig. 1 and Supplementary Fig. 1). We suspected that FSGS in both families was caused by a defect in the same gene. Under this assumption, the critical region for study was defined by the flanking SNPs rs3783397 (at approximately 103 Mb on the Marshfield map) and rs6576201 (at approximately 106 Mb).
We used public databases to identify genes in this region. We sequenced 15 genes in affected members of the FGBR and FGJN families. In each family, we found a sequence variant within the same exon of INF2 that segregated with disease and predicted a nonconservative amino-acid change (R218Q in FGJN and S186P in FGBR). We observed no likely disease-causing sequence variants in the other genes that were analyzed. The R218Q-encoding variant found in family FGJN was a de novo mutation, as the first individual in the family carrying this variant shared the disease-associated haplotype but not this variant with several unaffected siblings (data not shown).
We then sequenced INF2 in 91 unrelated individuals with familial FSGS. In probands from nine additional families (Supplementary Fig. 2), we identified point mutations in INF2 leading to nonconservative amino acid changes (Table 1, Fig. 2 and Supplementary Fig. 3). We typed these INF2 variants in available family members. We considered a family member to be affected if he or she had biopsy-confirmed FSGS, end-stage renal disease (ESRD) without other apparent cause, or substantial albuminuria (>250 mg albumin per gram of creatinine) without other apparent cause. We found that these mutations segregated with disease in each respective family (Fig. 1 and Supplementary Fig. 2). In five families, some younger individuals carrying these point mutations had no increase in urine protein, consistent with reduced, age-related penetrance, similar to the phenotypes associated with TRPC6 and ACTN4 mutations1,2,3. We found nucleotide variants in exons 8, 18 and 20 of INF2, but these did not segregate with disease and were also found in unaffected (control) individuals. All of the disease-segregating substitutions are located within the region of INF2 known as the diaphanous-inhibitory domain (DID)4 (Fig. 2b), and most of the mutations that result in these substitutions reside within exon 4 of INF2.
(a) Disease-segregating alterations shown aligned with wild-type INF2 protein sequence from humans, chimpanzee, mouse, rat, opossum and zebrafish. All of the disease alterations occur in evolutionarily conserved residues within the DID. (b) Schematic showing INF2 protein domain structure and location of the substitutions. (c–e) Model of mouse INF2 amino acids 1–330, based on the structure of mDia1. Residue variants are shown in red; residues important for the interaction with DAD are shown in blue. (c) View of mDia1 showing the positions of Ala13 and Arg218 (red). Residues important for the direct interaction with DAD are shown in blue, including Arg106 (corresponding to Lys213 in mDia1), Asn110 (corresponding to Asn217 in mDia1), Ala149 (corresponding to Ala256 in mDia1) and Ile152 (corresponding to Ile259 in mDia1). Based on the crystal structure of the mDia1 DID-DAD complex, the α-helical INF2 DAD is predicted to lie in the pocket containing these residues, with its N terminus (Asp974) contacting Arg106 and Asn110 and with Leu986 contacting Ala149 and Ile152. In this model, we predict that Arg218 would contact residues C-terminal to Leu986. (d) Close-up of the portion of the INF2 region predicted to interact with the DAD (shown with single-letter amino acid code). (e) 180-degree rotation of the structure shown in c, showing Leu42, Ser186 and Glu220.
To be certain that these variants were not polymorphisms and that INF2 is not a site of frequent but biologically insignificant variation, we resequenced exon 4, the location of all but two of the putative mutations, in 282 control individuals. None of these individuals carried any of these putative disease-causing INF2 variants or any other missense or splice-site variants. We genotyped the two putative disease-causing mutations found in exon 2 of INF2 (resulting in the A13T and L42P amino acid substitutions) as well as the mutations causing the E184K and R218Q substitutions, in an additional 341 control individuals using Sequenom assays. None of these variants were present in any of the 682 chromosomes assayed.
The phenotypes in families with INF2 mutations share certain features. Unlike the early-onset, nephrotic presentation seen in individuals with mutations in NPHS1 and NPHS2 (encoding the slit-diaphragm proteins nephrin and PODOCIN), individuals with INF2 mutations present in early adolescence or adulthood, typically with moderate proteinuria. Although we documented nephrotic-range proteinuria in members of several families, none of the affected individuals showed the spectrum of clinical findings that constitutes the so-called nephrotic syndrome. Microscopic hematuria and hypertension were noted in some individuals with INF2 mutations. Similar to what is seen in individuals with mutations in ACTN4, both disease and proteinuria were progressive, often leading to ESRD.
We reviewed available renal biopsy tissue samples from individuals with INF2 mutations (Fig. 3). Light microscopy typically showed the presence of FSGS (Fig. 3a), and electron microscopy showed focal areas of podocyte foot-process effacement, typical of secondary and some genetic forms of FSGS, as well as areas where foot processes and slit-diaphragms were well preserved. We also noted unusually prominent actin bundles within the foot processes (Fig. 3b). Glomerular hypertrophy was not a prominent feature of the biopsies. Neither we nor the original pathology reports noted the perihilar, collapsing or cellular lesions that characterize some subtypes of FSGS. Thus, these biopsies would be categorized as 'FSGS, not otherwise specified' following one widely adopted classification scheme5.
(a,b) Kidney biopsy findings in an affected member of family FSB with an E220K INF2 substitution. This individual was 21 years of age at the time of biopsy with an estimated glomerular filtration rate (GFR) of 96 ml/min/1.73 m2. She had 3+ urine protein and no hematuria. She developed ESRD 7 years after the biopsy was performed. Light micrograph (periodic acid–Schiff stain; a) shows focal global and segmental glomerulosclerosis. Electron micrograph (b) shows segmental foot-process effacement in some loops (arrowheads) and focally irregular morphology of preserved foot processes. Inset is a higher-magnification electron micrograph showing foot processes en face, projecting from a major process. The foot processes appear irregular and jagged, often with prominent longitudinal actin bundles. (c,d) Kidney biopsy findings from an individual from family FGJN with an R218Q substitution. This individual was 26 years of age at the time of biopsy. Estimated GFR was 46 ml/min/1.73 m2, and urine showed 3+ protein and trace blood at the time. Light micrograph (periodic acid–Schiff stain; c) shows focal and segmental glomerulosclerosis, with moderate chronic parenchymal damage. Electron micrograph (d) shows segmental foot-process effacement (arrowheads).
We examined the expression of INF2 in the kidney by in situ hybridization and by antibody staining (Fig. 4a,b). Both methods showed INF2 expression in the podocyte. We examined expression of INF2 and the slit-diaphragm protein nephrin together (Fig. 4b). INF2 expression showed areas of colocalization with nephrin in podocytes. Some INF2 staining was also noted in a pericapillary pattern, suggesting expression in cells other than podocytes. Within the podocyte, antibody staining showed what appeared to be a predominantly perinuclear pattern of INF2. RNA blot analysis showed INF2 expression in all tissues tested and identified INF2 transcripts of approximately 1.5 kb and 4.5 kb in length (Fig. 4c). This was consistent with previous reports describing INF2 and with the major transcripts predicted from ESTs and genomic sequence analysis as collated in the University of California at Santa Cruz (UCSC) Genome Browser4,6,7.
(a) RNA in situ hybridization in adult human kidney with digoxigenin-labeled probe targeted against INF2 mRNA, followed by immunohistochemical staining with labeled antibody to digoxigenin. INF2 mRNA expression is apparent in podocytes as well as some tubule cells. Sense control experiment is shown on the right. (b) Immunofluoresence staining of mouse kidney with the fluorescent stain DAPI (blue) and antibodies against nephrin (red) and INF2 (green). INF2 staining is observed in the glomeruli in an epithelial cell pattern. Arrows show areas of nephrin and INF2 localization. Arrowhead shows a cell expressing INF2 but not nephrin. (d) RNA blot. Lanes: 1, brain; 2, placenta; 3, skeletal muscle; 4, heart; 5, kidney; 6, pancreas; 7, liver; 8, lung; 9, spleen; 10, colon.
INF2 is a member of the formin family of actin-regulating proteins that accelerate actin polymerization8. All of the FSGS-associated sequence variants we identified lie within the DID. The DID is an autoinhibitory domain found in the diaphanous formins. In the most extensively studied diaphanous formin, mDia1, the interaction of the N-terminal DID with the C-terminal diaphanous activating domain (DAD) inhibits mDia1 function; this inhibition is relieved by the binding of mDia1 to Rho9. We used structural data available for the N-terminal region of mDia1 to map the altered INF2 residues and make functional predictions. Our INF2 model derived from the mDia1-RhoC structure (Fig. 2c–e) was in agreement with models derived from the other available mDia1 structures (data not shown)10. Two altered residues, Arg214 and Arg218, lie close to the DAD-binding region of the DID. Arg214 is on the surface of the DID in proximity to this binding site (Fig. 2c). Arg218 is predominately buried under the surface of the DID but has a small area of surface exposure in this region (Fig. 2d). Three other altered residues (Leu42, Ser186 and Glu220) are surface-exposed but on the opposite side of the DID structure from the DAD-binding region (Fig. 2e). The Ala13 position is surface-exposed and lies at the N terminus of the DID in the region corresponding to the GTPase binding domain of mDia1. Two of the altered residues are buried in the core DID structure (Glu184 and Leu198).
Based on these models, we can postulate potential functions for the variant INF2 residues. Arg214 and Arg218 likely form part of the DAD-binding site, given their spatial proximity to the key DAD-binding residues Ala149 and Ile152. The prominent exposure of Leu42, Ser186 and Glu220 on the DID surface raises the possibility that they interact with other molecules. Because the N terminus of mDia1 is important in the protein's cellular localization11, the N terminus of INF2 could also mediate localization. We postulate that Glu184, Leu198 and Arg218 are likely to be important for maintaining the overall fold of the DID. These residues are buried in the interior of the DID in all of our models, and the identified mutants (E184K, L198R and R218Q) could have effects on the integrity of the DID.
We engineered two substitutions (E184K and R218Q) predicted to have large effects on the structural integrity of the DID into the INF2 complementary DNA (cDNA). We also introduced a third mutation resulting in a substitution located away from the DAD binding site (S186P). These three alterations included the two (E186P and R218Q) found in the two large families (FGBR and FGJN) used in the initial genetic linkage analysis. We examined cultured podocytes transfected with wild-type or mutant INF2 constructs (resulting in the E184K, S186P and R218Q substitutions; Fig. 5). Podocytes transfected with the wild-type construct showed perinuclear INF2 staining, whereas podocytes transfected with INF2 harboring the E184K or R218Q alterations showed a different localization pattern, with a finer, more diffuse distribution. The cellular localization of the S186P INF2 mutant was closer to the pattern observed in the wild-type cells but had a more vermiform appearance. Both the wild-type and mutant forms of INF2 showed substantial co-staining with phalloidin, consistent with the notion that these proteins induced actin polymerization at their localization, and the overall staining of phalloidin mimicked that of INF2. In the mutant INF2-transfected cells, stress fibers and cortical actin were less prominent than in normal INF2-transfected cells. The similar and more marked changes seen with the E184K and R218Q mutants are consistent with their predicted effects on the overall INF2 DID structure. Furthermore, these data suggest that the DID is critical for the subcellular localization of INF2 and are consistent with data demonstrating that the DID is not required in order for INF2 to mediate actin polymerization7. Although these mutant forms of INF2 clearly behave differently than the wild-type protein, the relationship between this altered behavior and the disease mechanism remains to be clarified.
Images show localization of INF2 variants in undifferentiated cultured human podocytes. Cells were cultured on coverslips coated with type I collagen. INF2 is visualized using a Cy3-conjugated antibody against its C terminus7. Fluorescein isothiocyanate phalloidin was used to stain F-actin; nuclei are stained with DAPI. First column: cells expressing wild-type and three different mutant forms of INF2. Second column: INF2 channel. Third column: phalloidin. Rows: results for wild-type and three mutant forms of INF2 (E184K, S186P and R218Q). E184K and R218Q mutants show diffuse localization of INF2 and F-actin compared with the perinuclear INF2 staining seen with the wild-type expressing cells. Cells expressing the S186P mutant looked similar to wild-type–expressing cells, but INF2 expression was more vermiform in appearance. In all three mutants, stress fibers and cortical actin appeared to be less prominent than in wild-type cells.
The cumulative evidence we have presented shows that mutations in INF2 cause an autosomal dominant form of FSGS. In two large families, we found evidence of genetic linkage of the disease to a region on chromosome 14 containing INF2. We identified missense mutations in INF2 in 11 of 93 families examined. These mutations cause nonconservative substitutions in amino acids that are highly conserved, and the mutations segregate with disease within the affected families. Seven of the nine distinct mutations occur in close proximity to each other within exon 4 of INF2, and all nine mutations cause alterations within the DID. By resequencing exon 4 of INF2, we verified that neither these mutations nor other nonsynonymous DID variants are present in control groups. INF2 is highly expressed in the kidney, including in glomerular podocytes, the cell type thought to initiate most forms of FSGS. Electron microscopy of kidney biopsy material from an individual with an INF2 mutation showed irregular podocyte foot-process morphology and prominent actin bundles. In addition, transfection studies of overexpressed wild-type and FSGS-associated mutant forms of INF2 indicate differences in the subcellular localization of the mutant and wild-type proteins, as well as differences in the pattern of distribution of the associated F-actin. In the electron micrographs, prominent actin filament bundles are noted, whereas actin filaments appear somewhat less prominent than normal in the INF2-mutant–expressing cells. The precise mechanism by which actin behavior is disrupted in the presence of INF2 alterations in vivo remains to be defined.
Formins are a family of proteins that accelerate actin filament assembly12. INF2 has the unique ability to accelerate both polymerization and depolymerization of actin in vitro, and it localizes to the endoplasmic reticulum (ER) in fibroblasts. All of the disease-causing INF2 mutations we identified affect the N-terminal regulatory region of the protein containing the DID7. The DID binds the DAD region of INF2 and autoinhibits INF2-mediated actin depolymerization but does not inhibit actin polymerization7. Previously published work shows that expression of an INF2 mutant in which the DID-DAD interaction is blocked causes actin filament accumulation around the ER and collapse of the ER onto the nuclear membrane7. Here we find that substitutions in the DID predicted to severely affect the domain structure (E184K and R218Q) do not appear to prevent INF2-mediated actin filament accumulation but do alter INF2 and F-actin localization.
Podocytes are complex, actin-rich, interdigitating structures that must be able to react to a unique set of physical and chemical stresses. Dysregulation of the podocyte cytoskeleton is a common feature of glomerular disease states13. The proximal trigger leading to altered actin behavior in podocytes can derive from external signals (for example, from the slit-diaphragm14) or internally (as seen in ACTN4 mutations1). Similar to what has been observed with α-actinin-4, INF2 is a widely expressed actin-binding protein that, when mutated, causes a podocyte-limited phenotype. Our results support a model of podocytes as dynamic structures that are extremely sensitive to alterations in the spatial or temporal regulation of the actin cytoskeleton. We hypothesize that individuals harboring disease-associated INF2 mutations have a defect in actin-mediated podocyte structural maintenance and repair. Future studies aimed at defining how INF2 mutations lead to disease will contribute to our understanding of podocyte biology and FSGS, as well as common forms of kidney disease characterized by podocyte dysfunction.
Methods
Clinical ascertainment.
Subjects were enrolled in these studies after we obtained informed consent in accordance with a human subjects protocol approved by the Brigham and Women's Hospital. We extracted DNA from peripheral blood, measured urine microalbumin and creatinine concentrations and obtained clinical and family history information. We reviewed medical records, kidney biopsies and biopsy reports when available. Additional details are provided in the Supplementary Note.
Genotyping.
In family FGBR, we performed a genome-wide analysis using DNA samples from 31 informative family members and approximately 300 microsatellite markers from the Marshfield map using standard methodology. We computed parametric two-point and multipoint log likelihood ratio scores using the FASTLINK15 and VITESSE programs16. In family FGJN, we performed genome-wide linkage analysis using 250K Affymetrix SNP Chips and follow-up genotyping using 14q microsatellite markers, and we performed parametric multipoint linkage analysis using both LINKAGE and GENEHUNTER17. Additional genotyping of the identified variants was performed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Sequenom) at the Harvard-Partners Core Genotyping Facility.
Sequencing.
Using standard Sanger sequencing methodology on an Applied Biosystems 3730 DNA Analyzer, we sequenced PCR-amplified segments containing coding sequences and flanking splice sites of 15 genes (INF2, ADSSL1, SIVA1, AKT1, PLD4, AHNAK2, CDCA4, GPR132, JAG2, NUDT14, BRF1, PACS2, MTA1, CRIP2 and CRIP1) from the 14q critical region in 12 individuals (3 affected and 3 unaffected members each from both family FGBR and family FGJN) and then carried out additional sequencing of INF2 in family members and control individuals. Sequencing primers are available on request.
Structural model.
We designed structural models of INF2 using the Phyre resource from Imperial College, London18. We submitted amino acids 1–424 of mouse INF2 and recovered INF2 models based on the structural coordinates of three mDia1 N-terminal structures10,19,20. (The fourth mDia1 N-terminal structure21 was not represented in this search.) We manipulated the models using the program PyMOL. In the model, the identified FSGS-associated alterations correspond to residues in mDia1 in the structural alignment (Supplementary Table 1).
INF2 mutagenesis.
We obtained a clone containing full-length INF2 in the pCMV6-XL5 vector (Origene). Mutagenesis was carried out using the Stratagene QuikChange II kit and mutagenesis primers designed to create the E184K, S186P and R218Q variants. All mutagenesis reactions were verified by sequencing.
INF2 expression.
For RNA blotting, a human multiple tissue RNA blot was purchased from Ambion. 5′ sense and antisense probes consisting of the first 590 bp of INF2 were made by [α-32P]dCTP labeling of a PCR-generated fragment using Klenow enzyme (New England Biolabs NEBlot kit). For in situ hybridization, a nonradioactive in situ hybridization was performed as described22, using a digoxigenin (DIG)-labeled cRNA probe that contained the first 590 bases of the INF2 sequence. Frozen sections (10 μm) of human kidney tissue were cut in a cryostat and captured onto Superfrost plus microscope slides (Fisher Scientific). Sections were then fixed and acetylated and hybridized to the probe at 70 °C for approximately 72 h (approximate concentration 100 ng/ml). The hybridized probe was visualized using alkaline phosphatase–conjugated anti-DIG Fab fragments (Roche) and 5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium (BCIP/NBT) substrate (Kierkegard and Perry Laboratories). Sections were rinsed several times in 100 mM Tris, 150 mM sodium chloride, 20 mM EDTA at pH 9.5 and coverslipped with glycerol gelatin (Sigma). Control sections were incubated in an identical concentration of the sense probe transcript.
Microscopy studies.
Undifferentiated human podocytes in culture (gift of M. Saleem, University of Bristol, UK) were transfected with wild-type or mutant INF2 constructs using FuGENE (Roche). We used antibody to the C terminus of INF2 directly conjugated to Cyanine (Cy)3 at 1:200 dilution to visualize wild-type and mutant INF2 protein7. We used fluorescein isothiocyanate phalloidin and the fluorescent stain DAPI to visualize actin filaments and nuclei.
For kidney sections, we made 5-μm sections of acetone-fixed kidneys from healthy 6-week-old mice. After washing with PBS, we incubated sections with primary antibodies: rabbit anti-INF2 (C-terminal 300 amino acids)7 and guinea pig anti-nephrin (Progen Biotechnik). We used anti-rabbit Cy2-coupled and anti-guinea pig Cy3-coupled secondary antibodies to visualize INF2 and nephrin, and DAPI to visualize nuclei. We analyzed specimens using a Nikon TE-200E inverted microscope and AutoDeblur deconvolution software (Media Cybernetics).
URLs.
PyMOL software, http://www.pymol.org/.
Change history
05 February 2010
In the version of this article initially published, Stephen J. Tonna was inadvertently omitted from the author list, and the fourth author (Hiroyasu Tsukaguchi) was missing one of his affiliations. These errors have been corrected in the HTML and PDF versions of the article.
References
Kaplan, J.M. et al. Mutations in ACTN4, encoding α-actinin-4, cause familial focal segmental glomerulosclerosis. Nat. Genet. 24, 251–256 (2000).
Winn, M.P. et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science 308, 1801–1804 (2005).
Reiser, J. et al. TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat. Genet. 37, 739–744 (2005).
Chhabra, E.S. & Higgs, H.N. INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 281, 26754–26767 (2006).
D'Agati, V.D., Fogo, A.B., Bruijn, J.A. & Jennette, J.C. Pathologic classification of focal segmental glomerulosclerosis: a working proposal. Am. J. Kidney Dis. 43, 368–382 (2004).
Kent, W.J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Chhabra, E.S., Ramabhadran, V., Gerber, S.A. & Higgs, H.N. INF2 is an endoplasmic reticulum-associated formin protein. J. Cell Sci. 122, 1430–1440 (2009).
Faix, J. & Grosse, R. Staying in shape with formins. Dev. Cell 10, 693–706 (2006).
Li, F. & Higgs, H.N. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 (2003).
Rose, R. et al. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 435, 513–518 (2005).
Seth, A., Otomo, C. & Rosen, M.K. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLα and mDia1. J. Cell Biol. 174, 701–713 (2006).
Higgs, H.N. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30, 342–353 (2005).
Faul, C., Asanuma, K., Yanagida-Asanuma, E., Kim, K. & Mundel, P. Actin up: regulation of podocyte structure and function by components of the actin cytoskeleton. Trends Cell Biol. 17, 428–437 (2007).
Huber, T.B. & Benzing, T. The slit diaphragm: a signaling platform to regulate podocyte function. Curr. Opin. Nephrol. Hypertens. 14, 211–216 (2005).
Schäffer, A.A. Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum. Hered. 46, 226–235 (1996).
O'Connell, J.R. & Weeks, D.E. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat. Genet. 11, 402–408 (1995).
Markianos, K., Daly, M.J. & Kruglyak, L. Efficient multipoint linkage analysis through reduction of inheritance space. Am. J. Hum. Genet. 68, 963–977 (2001).
Kelley, L.A. & Sternberg, M.J. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009).
Otomo, T. et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–494 (2005).
Lammers, M., Rose, R., Scrima, A. & Wittinghofer, A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 24, 4176–4187 (2005).
Nezami, A.G., Poy, F. & Eck, M.J. Structure of the autoinhibitory switch in formin mDia1. Structure 14, 257–263 (2006).
Berger, U.V. & Hediger, M.A. Differential distribution of the glutamate transporters GLT-1 and GLAST in tanycytes of the third ventricle. J. Comp. Neurol. 433, 101–114 (2001).
Acknowledgements
We thank the many individuals who participated in this study; S. DePalma for help with linkage analysis; and J. Jacobs, K. Tucker, J. Holt, A. Acharya, R.Wiggins, R.Weir, J. Levine and many others for help with family ascertainment. This work was support by grants from the US National Institutes of Health (DK54931 to M.R.P., DK073091 to J.M.H., DK080947 to J.S.S. and GM069818 to H.N.H.) and the Clinical Investigator Training Program: Beth Israel Deaconess Medical School in collaboration with Pfizer Inc. and Merck and Co., the NephCure Foundation, and the Cole Pasqualucci Nephrotic Syndrome and FSGS Research fund (to E.J.B.). M.R.P. is an Established Investigator of the American Heart Association.
Author information
Authors and Affiliations
Contributions
E.J.B. performed family and clinical ascertainment, performed and interpreted genetic linkage studies, performed mutational analysis and made DNA constructs. J.S.S. helped with study design and interpretation and performed INF2 expression studies. D.J.B. performed INF2 expression studies. H.T. performed genetic linkage studies. S.J.T. performed genotyping and sequencing analyses. A.L.U. helped with clinical ascertainment of the families. H.N.H. performed molecular modeling and assisted with data interpretation. J.M.H. performed histology studies. M.R.P. oversaw all aspects of study design and interpretation and wrote the manuscript with the assistance of the other authors.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1 and Supplementary Note (PDF 781 kb)
Rights and permissions
About this article
Cite this article
Brown, E., Schlöndorff, J., Becker, D. et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 42, 72–76 (2010). https://doi.org/10.1038/ng.505
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.505
This article is cited by
-
Phosphorylation of INF2 by AMPK promotes mitochondrial fission and oncogenic function in endometrial cancer
Cell Death & Disease (2024)
-
Phenotype-driven reanalysis reveals five novel pathogenic variants in 40 exome-negative families with Charcot–Marie–Tooth Disease
Journal of Neurology (2024)
-
Hidden genetics behind glomerular scars: an opportunity to understand the heterogeneity of focal segmental glomerulosclerosis?
Pediatric Nephrology (2023)
-
Structure and function of the N-terminal extension of the formin INF2
Cellular and Molecular Life Sciences (2022)
-
Causal and putative pathogenic mutations identified in 39% of children with primary steroid-resistant nephrotic syndrome in South Africa
European Journal of Pediatrics (2022)