Abstract
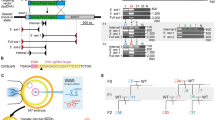
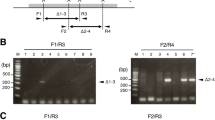
Here we describe a practical Cre-loxP and piggyBac transposon–based mutagenesis strategy to systematically mutate coding sequences and/or the vast noncoding regions of the mouse genome for large-scale functional genomic analysis. To illustrate this approach, we first created loxP-containing loss-of-function alleles in the protocadherin α, β and γ gene clusters (Pcdha, Pcdhb and Pcdhg). Using these alleles, we show that, under proper guidance, Cre-loxP site-specific recombination can mediate efficient trans-allelic recombination in vivo, facilitating the generation of large germline deletions and duplications including deletions of Pcdha, and Pcdha to Pcdhb, simply by breeding (that is, at frequencies of 5.5%–21.6%). The same breeding method can also generate designed germline translocations between nonhomologous chromosomes at unexpected frequencies of greater than 1%. By incorporating a piggyBac transposon to insert and to distribute loxP sites randomly throughout the mouse genome, we present a simple but comprehensive method for generating genome-wide deletions and duplications, in addition to insertional loss-of-function and conditional rescue alleles, again simply by breeding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Austin, C.P. et al. The knockout mouse project. Nat. Genet. 36, 921–924 (2004).
Auwerx, J. et al. The European dimension for the mouse genome mutagenesis program. Nat. Genet. 36, 925–927 (2004).
The International Mouse Knockout Consortium. A mouse for all reasons. Cell 128, 9–13 (2007).
Dermitzakis, E.T., Reymond, A. & Antonarakis, S.E. Conserved non-genic sequences — an unexpected feature of mammalian genomes. Nat. Rev. Genet. 6, 151–157 (2005).
Bejerano, G. et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004).
Boffelli, D., Nobrega, M.A. & Rubin, E.M. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 5, 456–465 (2004).
Sandelin, A. et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5, 99 (2004).
Margulies, E.H. et al. Comparative sequencing provides insights about the structure and conservation of marsupial and monotreme genomes. Proc. Natl. Acad. Sci. USA 102, 3354–3359 (2005).
Vavouri, T., McEwen, G.K., Woolfe, A., Gilks, W.R. & Elgar, G. Defining a genomic radius for long-range enhancer action: duplicated conserved non-coding elements hold the key. Trends Genet. 22, 5–10 (2006).
Bejerano, G., Haussler, D. & Blanchette, M. Into the heart of darkness: large-scale clustering of human non-coding DNA. Bioinformatics 20, I40–I48 (2004).
Golic, K.G. & Golic, M.M. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144, 1693–1711 (1996).
Ryder, E. et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167, 797–813 (2004).
Zheng, B., Sage, M., Sheppeard, E.A., Jurecic, V. & Bradley, A. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol. Cell. Biol. 20, 648–655 (2000).
Mills, A.A. & Bradley, A. From mouse to man: generating megabase chromosome rearrangements. Trends Genet. 17, 331–339 (2001).
Herault, Y., Rassoulzadegan, M., Cuzin, F. & Duboule, D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE). Nat. Genet. 20, 381–384 (1998).
Kmita, M., Fraudeau, N., Herault, Y. & Duboule, D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 420, 145–150 (2002).
Zakany, J. & Duboule, D. Hox genes and the making of sphincters. Nature 401, 761–762 (1999).
Genoud, N. et al. Disruption of Doppel prevents neurodegeneration in mice with extensive Prnp deletions. Proc. Natl. Acad. Sci. USA 101, 4198–4203 (2004).
Spitz, F., Herkenne, C., Morris, M.A. & Duboule, D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat. Genet. 37, 889–893 (2005).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Wu, Q. & Maniatis, T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790 (1999).
Wu, Q. et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11, 389–404 (2001).
Wu, Q. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics 169, 2179–2188 (2005).
Kohmura, N. et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20, 1137–1151 (1998).
Wang, X. et al. Gamma protocadherins are required for survival of spinal interneurons. Neuron 36, 843–854 (2002).
Phillips, G.R. et al. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 23, 5096–5104 (2003).
Weiner, J.A., Wang, X., Tapia, J.C. & Sanes, J.R. Gamma protocadherins are required for synaptic development in the spinal cord. Proc. Natl. Acad. Sci. USA 102, 8–14 (2005).
Hambsch, B., Grinevich, V., Seeburg, P.H. & Schwarz, M.K. γ-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. J. Biol. Chem. 280, 15888–15897 (2005).
Tang, S.H., Silva, F.J., Tsark, W.M. & Mann, J.R. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32, 199–202 (2002).
Bonin, C.P. & Mann, R.S. A piggyBac transposon gene trap for the analysis of gene expression and function in Drosophila. Genetics 167, 1801–1811 (2004).
Thibault, S.T. et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283–287 (2004).
Schmidt, E.E., Taylor, D.S., Prigge, J.R., Barnett, S. & Capecchi, M.R. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl. Acad. Sci. USA 97, 13702–13707 (2000).
DeChiara, T.M. et al. Ror2, encoding a receptor-like tyrosine kinase, is required for cartilage and growth plate development. Nat. Genet. 24, 271–274 (2000).
Komori, T. et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 (1997).
Jung, J.Y. et al. Expression of urea transporters in potassium-depleted mouse kidney. Am. J. Physiol. Renal Physiol. 285, F1210–F1224 (2003).
Forster, A. et al. Engineering de novo reciprocal chromosomal translocations associated with Mll to replicate primary events of human cancer. Cancer Cell 3, 449–458 (2003).
Zong, H., Espinosa, J.S., Su, H.H., Muzumdar, M.D. & Luo, L. Mosaic analysis with double markers in mice. Cell 121, 479–492 (2005).
Dracopoli, N.C. et al. in Current Protocols in Human Genetics 4.1.1–4.1.4 (John Wiley & Sons, Hoboken, New Jersey, 2006).
Cary, L.C. et al. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172, 156–169 (1989).
Fraser, M.J., Ciszczon, T., Elick, T. & Bauser, C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 5, 141–151 (1996).
Li, X. et al. piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol. Biol. 14, 17–30 (2005).
Friedrich, G. & Soriano, P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5, 1513–1523 (1991).
Liu, P., Jenkins, N.A. & Copeland, N.G. Efficient Cre-loxP-induced mitotic recombination in mouse embryonic stem cells. Nat. Genet. 30, 66–72 (2002).
Handler, A.M. & Harrell, R.A. II Transformation of the Caribbean fruit fly, Anastrepha suspensa, with a piggyBac vector marked with polyubiquitin-regulated GFP. Insect Biochem. Mol. Biol. 31, 199–205 (2001).
Boulet, A.M. & Capecchi, M.R. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development 131, 299–309 (2004).
Economides, K.D., Zeltser, L. & Capecchi, M.R. Hoxb13 mutations cause overgrowth of caudal spinal cord and tail vertebrae. Dev. Biol. 256, 317–330 (2003).
Bunting, M., Bernstein, K.E., Greer, J.M., Capecchi, M.R. & Thomas, K.R. Targeting genes for self-excision in the germ line. Genes Dev. 13, 1524–1528 (1999).
Acknowledgements
We thank J.R. Mann (City of Hope's Beckman Research Institute) for the Hprt-Cre mouse; J.-F. Cheng (Lawrence Berkeley National Laboratory) for the Pcdh BAC clones; C. Zou and H. Peng (laboratory of Q.W.) for constructing Pcdha1 and Pcdhga1 targeting vectors and performing DNA blot experiments; M. Hockin for help with chromosome painting; G. Karan for fluorescence photography of adult mice; members of M.R.C.'s laboratory for comments on the manuscript and the University of Utah Transgenic/Gene Targeting Facility for pronuclear injections. pBigT was a gift of F. Constantini (Columbia University), plasmid C4-PBss was gift of R.S. Mann (Columbia University) and plasmid 286 was a gift of A. Handler (University of Florida). We are grateful for technical support from the ES cell culture, mouse surgery and husbandry staff in M.R.C.'s laboratory, in particular S. Barnett, L. Byers, C. Lenz, K. Lustig, J. Tomlin and J. Shuhua. G.Y. and Q.W. were supported by a grant from the American Cancer Society. Q.W. is a Basil O'Connor Scholar.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed by the University of Utah that includes some of the work presented in this publication.
Supplementary information
Supplementary Fig. 1
Gene targeting strategy for the Pcdha1eGFP, Pcdhac1AP, Pcdhac2lacZ, PcdhaCon and Pcdhga1hrGFP knockout alleles. (PDF 224 kb)
Supplementary Fig. 2
Normal gross anatomy in the brain of an adult del(α)/del(α) mouse. (PDF 797 kb)
Supplementary Fig. 3
Variations of ZG-s gene-trap vectors. (PDF 313 kb)
Supplementary Fig. 4
X-gal staining of more examples of gene-trap alleles and somatic transallelic recombination using gene-trap loxP alleles. (PDF 277 kb)
Supplementary Table 1
Genotyping primers for all alleles in this study. (PDF 101 kb)
Supplementary Table 2
piggyBac transpositions in founder mice. (PDF 17 kb)
Supplementary Table 3
piggyBac transpositions in the germline through breeding. (PDF 20 kb)
Rights and permissions
About this article
Cite this article
Wu, S., Ying, G., Wu, Q. et al. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet 39, 922–930 (2007). https://doi.org/10.1038/ng2060
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng2060
This article is cited by
-
Murine allele and transgene symbols: ensuring unique, concise, and informative nomenclature
Mammalian Genome (2022)
-
Cas12a mediates efficient and precise endogenous gene tagging via MITI: microhomology-dependent targeted integrations
Cellular and Molecular Life Sciences (2020)
-
Comparing the utility of in vivo transposon mutagenesis approaches in yeast species to infer gene essentiality
Current Genetics (2020)
-
New Approaches in Cognitive Neurobiology: Methods of Molecular Marking and Ex Vivo Imaging of Cognitively Active Neurons
Neuroscience and Behavioral Physiology (2018)
-
Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE
Scientific Reports (2017)