Abstract
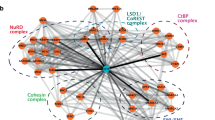
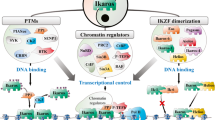
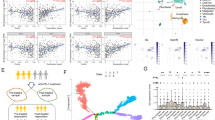
Cell fate depends on the interplay between chromatin regulators and transcription factors. Here we show that activity of the Mi-2β nucleosome-remodeling and histone-deacetylase (NuRD) complex was controlled by the Ikaros family of lymphoid lineage–determining proteins. Ikaros, an integral component of the NuRD complex in lymphocytes, tethered this complex to active genes encoding molecules involved in lymphoid differentiation. Loss of Ikaros DNA-binding activity caused a local increase in chromatin remodeling and histone deacetylation and suppression of lymphoid cell–specific gene expression. Without Ikaros, the NuRD complex also redistributed to transcriptionally poised genes that were not targets of Ikaros (encoding molecules involved in proliferation and metabolism), which induced their reactivation. Thus, release of NuRD from Ikaros regulation blocks lymphocyte maturation and mediates progression to a leukemic state by engaging functionally opposing epigenetic and genetic networks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Roh, T.Y., Cuddapah, S. & Zhao, K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19, 542–552 (2005).
Li, B., Carey, M. & Workman, J.L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Wang, Z. et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009).
Tong, J.K., Hassig, C.A., Schnitzler, G.R., Kingston, R.E. & Schreiber, S.L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 (1998).
Zhang, Y., LeRoy, G., Seelig, H.P., Lane, W.S. & Reinberg, D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 (1998).
Ho, L. & Crabtree, G.R. Chromatin remodelling during development. Nature 463, 474–484 (2010).
Kim, J. et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10, 345–355 (1999).
O'Neill, D.W. et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol. 20, 7572–7582 (2000).
Georgopoulos, K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat. Rev. Immunol. 2, 162–174 (2002).
Yoshida, T., Ng, S.Y., Zuniga-Pflucker, J.C. & Georgopoulos, K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 7, 382–391 (2006).
Ng, S.Y., Yoshida, T., Zhang, J. & Georgopoulos, K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30, 493–507 (2009).
Morgan, B. et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 16, 2004–2013 (1997).
Winandy, S., Wu, P. & Georgopoulos, K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83, 289–299 (1995).
Wang, J.H. et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5, 537–549 (1996).
Dumortier, A. et al. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol. Cell. Biol. 26, 209–220 (2006).
Gómez-del Arco, P. et al. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity 24, 685–698 (2010).
Williams, C.J. et al. The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity 20, 719–733 (2004).
Naito, T., Gomez-Del Arco, P., Williams, C.J. & Georgopoulos, K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2β determine silencer activity and Cd4 gene expression. Immunity 27, 723–734 (2007).
Avitahl, N. et al. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity 10, 333–343 (1999).
Singer, A., Adoro, S. & Park, J.H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8, 788–801 (2008).
Koipally, J. & Georgopoulos, K. A molecular dissection of the repression circuitry of Ikaros. J. Biol. Chem. 277, 27697–27705 (2002).
Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58, 236–242 (1963).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Roh, T.Y., Cuddapah, S., Cui, K. & Zhao, K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA 103, 15782–15787 (2006).
Bernstein, B.E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Molnár, A. & Georgopoulos, K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 14, 8292–8303 (1994).
Hahm, K. et al. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 12, 782–796 (1998).
Hollenhorst, P.C., Shah, A.A., Hopkins, C. & Graves, B.J. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21, 1882–1894 (2007).
Murre, C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6, 1079–1086 (2005).
Kee, B.L. E and ID proteins branch out. Nat. Rev. Immunol. 9, 175–184 (2009).
Jones, M.E. & Zhuang, Y. Stage-specific functions of E-proteins at the beta-selection and T-cell receptor checkpoints during thymocyte development. Immunol. Res. 49, 202–215 (2011).
Lin, Y.C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11, 635–643 (2010).
Gould, A. Functions of mammalian Polycomb group and trithorax group related genes. Curr. Opin. Genet. Dev. 7, 488–494 (1997).
Mullighan, C.G. et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–764 (2007).
Aichinger, E., Villar, C.B., Di Mambro, R., Sabatini, S. & Kohler, C. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23, 1047–1060 (2011).
Hakimi, M.A. et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418, 994–998 (2002).
Wendt, K.S. et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 (2008).
Kagey, M.H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Kaufmann, C. et al. A complex network of regulatory elements in Ikaros and their activity during hemo-lymphopoiesis. EMBO J. 22, 2211–2223 (2003).
Gómez-del Arco, P., Maki, K. & Georgopoulos, K. Phosphorylation controls Ikaros's ability to negatively regulate the G1-S transition. Mol. Cell. Biol. 24, 2797–2807 (2004).
Gómez-del Arco, P., Koipally, J. & Georgopoulos, K. Ikaros SUMOylation: switching out of repression. Mol. Cell. Biol. 25, 2688–2697 (2005).
Mavrakis, K.J. et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat. Genet. 43, 673–678 (2011).
Masetti, R., Serravalle, S., Biagi, C. & Pession, A. The role of HDACs inhibitors in childhood and adolescence acute leukemias. J. Biomed. Biotechnol. 2011, 148046 (2011).
Mullighan, C.G. et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471, 235–239 (2011).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1–25 (2004).
Griffith, A.V. et al. Spatial mapping of thymic stromal microenvironments reveals unique features influencing T lymphoid differentiation. Immunity 31, 999–1009 (2009).
Acknowledgements
We thank P. Gomez for analysis of the expression of Mi-2β protein in thymocytes; the Kingston laboratory (Massachusetts General Hospital) for the hSWI-SNF complex and help with setting up the mononucleosome and 5S array assays; I. Joshi for help with cell sorting; B. Czyzewski for mouse husbandry; and B. Morgan for discussions of the project and critical review of the manuscript. Protein microsequencing was done at the Microchemistry and Proteomics Facility of Harvard University, Cambridge, and at the Taplin Mass Spectrometry Facility of Harvard Medical School, Boston, and high-throughput DNA sequencing and RNA profiling were done at the Bauer Center for Genomic Research of Harvard University, Cambridge. Supported by the US National Institutes of Health (2T32AI007529 to A.F.J.; and 5R01AI042254 and R01CA158006 to K.G.).
Author information
Authors and Affiliations
Contributions
J.Z., A.F.J., T.N., M.D., E.J.H., J.S., F.L., M.K. and T.Y. designed and did experiments and analyzed experimental data; H.T.P. designed expression studies of wild-type thymocyte subsets and provided data for analysis; F.G. and K.G. supervised research and analyzed data; and J.Z. and K.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1–2 and Methods (PDF 1068 kb)
Rights and permissions
About this article
Cite this article
Zhang, J., Jackson, A., Naito, T. et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13, 86–94 (2012). https://doi.org/10.1038/ni.2150
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2150
This article is cited by
-
The transcription factor Aiolos restrains the activation of intestinal intraepithelial lymphocytes
Nature Immunology (2024)
-
IKAROS: from chromatin organization to transcriptional elongation control
Cell Death & Differentiation (2023)
-
How transcription factors drive choice of the T cell fate
Nature Reviews Immunology (2021)
-
A variant in human AIOLOS impairs adaptive immunity by interfering with IKAROS
Nature Immunology (2021)
-
CHD4/NuRD complex regulates complement gene expression and correlates with CD8 T cell infiltration in human hepatocellular carcinoma
Clinical Epigenetics (2020)