Abstract
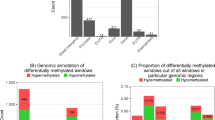
Beyond its well-characterized functions in antibody diversification, the cytidine deaminase AID can catalyze off-target DNA damage and has been hypothesized to edit RNA and mediate DNA demethylation. To comprehensively examine the effects of AID on the transcriptome and the pattern of DNA methylation ('methylome'), we analyzed AID-deficient (Aicda−/−), wild-type and AID-overexpressing activated B cells by high-throughput RNA sequencing (RNA-Seq) and reduced-representation bisulfite sequencing (RRBS). These analyses confirmed the known role of AID in immunoglobulin isotype switching and also demonstrated few other effects of AID on gene expression. Additionally, we detected no evidence of AID-dependent editing of mRNA or microRNA. Finally, the RRBS data did not support the proposed role for AID in regulating DNA methylation. Thus, despite evidence of its additional activities in other systems, antibody diversification seems to be the sole physiological function of AID in activated B cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000).
Revy, P. et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102, 565–575 (2000).
Gazumyan, A., Bothmer, A., Klein, I.A., Nussenzweig, M.C. & McBride, K.M. Activation-induced cytidine deaminase in antibody diversification and chromosome translocation. Adv. Cancer Res. 113, 167–190 (2012).
Pavri, R. & Nussenzweig, M.C. AID targeting in antibody diversity. Adv. Immunol. 110, 1–26 (2011).
Bransteitter, R., Pham, P., Scharff, M.D. & Goodman, M.F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA 100, 4102–4107 (2003).
Chaudhuri, J. et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730 (2003).
Dickerson, S.K., Market, E., Besmer, E. & Papavasiliou, F.N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197, 1291–1296 (2003).
Nonaka, T. et al. Carboxy-terminal domain of AID required for its mRNA complex formation in vivo. Proc. Natl. Acad. Sci. USA 106, 2747–2751 (2009).
Kobayashi, M. et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc. Natl. Acad. Sci. USA 106, 22375–22380 (2009).
Liang, G. et al. RNA editing of hepatitis B virus transcripts by activation-induced cytidine deaminase. Proc. Natl. Acad. Sci. USA 110, 2246–2251 (2013).
Wang, C.L., Harper, R.A. & Wabl, M. Genome-wide somatic hypermutation. Proc. Natl. Acad. Sci. USA 101, 7352–7356 (2004).
Liu, M. et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature 451, 841–845 (2008).
Shen, H.M., Peters, A., Baron, B., Zhu, X. & Storb, U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 280, 1750–1752 (1998).
Pasqualucci, L. et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl. Acad. Sci. USA 95, 11816–11821 (1998).
Ramiro, A.R. et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118, 431–438 (2004).
Robbiani, D.F. et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135, 1028–1038 (2008).
Robbiani, D.F. et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell 36, 631–641 (2009).
Klein, I.A. et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106 (2011).
Chiarle, R. et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 (2011).
Pasqualucci, L. et al. AID is required for germinal center–derived lymphomagenesis. Nat. Genet. 40, 108–112 (2007).
Yamane, A. et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 12, 62–69 (2010).
Morgan, H.D., Dean, W., Coker, H.A., Reik, W. & Petersen-Mahrt, S.K. Activation-induced cytidine deaminase deaminates 5-Methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 279, 52353–52360 (2004).
Bhutani, N. et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463, 1042–1047 (2010).
Rai, K. et al. DNA Demethylation in zebrafish Involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell 135, 1201–1212 (2008).
Popp, C. et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463, 1101–1105 (2010).
Bhutani, N. et al. A critical role for AID in the initiation of reprogramming to induced pluripotent stem cells. FASEB J. 27, 1107–1113 (2013).
Rai, K. et al. DNA Demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell 142, 930–942 (2010).
Cortellino, S. et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 (2011).
Fritz, E.L. & Papavasiliou, F.N. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 24, 2107–2114 (2010).
Guo, J.U., Su, Y., Zhong, C., Ming, G.-L. & Song, H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell 145, 423–434 (2011).
Nabel, C.S. et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 8, 751–758 (2012).
Wijesinghe, P. & Bhagwat, A.S. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 40, 9206–9217 (2012).
Teng, G. et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 28, 621–629 (2008).
Jiang, L. et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21, 1543–1551 (2011).
Meyers, G. et al. Activation-induced cytidine deaminase (AID) is required for B-cell tolerance in humans. Proc. Natl. Acad. Sci. USA 108, 11554–11559 (2011).
Kuraoka, M. et al. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc. Natl. Acad. Sci. USA 108, 11560–11565 (2011).
Rosenberg, B.R., Hamilton, C.E., Mwangi, M.M., Dewell, S. & Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 18, 230–236 (2011).
Kobayashi, M. et al. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc. Natl. Acad. Sci. USA 108, 19305–19310 (2011).
Yang, W. et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 13, 13–21 (2005).
Kawahara, Y. et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315, 1137–1140 (2007).
Alon, S. et al. Systematic identification of edited microRNAs in the human brain. Genome Res. 22, 1533–1540 (2012).
Vesely, C., Tauber, S., Sedlazeck, F.J., Haeseler, von, A. & Jantsch, M.F. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 22, 1468–1476 (2012).
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008).
Bock, C. et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol. Cell 47, 633–647 (2012).
Shaknovich, R. et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 118, 3559–3569 (2011).
Abdouni, H. et al. Zebrafish AID is capable of deaminating methylated deoxycytidines. Nucl. Acids Res. advance online publication, doi:10.1093/nar/gkt212 (12 April 2013).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Lefranc, M.-P. et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 37, D1006–D1012 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Hafner, M. et al. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods 58, 164–170 (2012).
Robinson, J.T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Krueger, F. & Andrews, S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Team, R.D.C. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2012).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Acknowledgements
We thank members of the F.N.P. laboratory for discussion; C. Hamilton and D. Schulz for comments on the manuscript; C. Mason (Weill Cornell Medical College) for ERCC spikes; G. Hovel-Miner for VSG constructs; K. Velinzon for cell sorting; S. Dewell for sequencing advice; and Y. Li for assistance with EpiTYPER assays. Supported by the National Cancer Institute (CA098495), the Starr Cancer Consortium (I4-A447), the Howard Hughes Medical Institute (T.T.) and the US National Institutes of Health (work in the T.T. laboratory).
Author information
Authors and Affiliations
Contributions
E.L.F., B.R.R. and F.N.P. designed experiments and analyses; E.L.F. and K.L. cultured cells; K.L. generated VSG spikes; A.M. prepared miRNA sequencing libraries with the supervision of T.T.; E.L.F. did all other experiments and analyzed the data; and E.L.F., B.R.R. and F.N.P. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 1567 kb)
Supplementary Table 1
Genes with greater than 2-fold difference between conditions, and full gene expression tables (XLSX 6595 kb)
Supplementary Table 2
Annotation of I-C and JH-C transcripts for quantification by Cufflinks (TXT 38 kb)
Supplementary Table 3
VH segment usage by AID expression, as determined by RNA-Seq (XLSX 67 kb)
Rights and permissions
About this article
Cite this article
Fritz, E., Rosenberg, B., Lay, K. et al. A comprehensive analysis of the effects of the deaminase AID on the transcriptome and methylome of activated B cells. Nat Immunol 14, 749–755 (2013). https://doi.org/10.1038/ni.2616
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2616
This article is cited by
-
Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination
Nature Reviews Genetics (2022)
-
Activation-induced cytidine deaminase targets SUV4-20-mediated histone H4K20 trimethylation to class-switch recombination sites
Scientific Reports (2017)
-
Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design
BMC Biotechnology (2016)
-
Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity
Nature Reviews Immunology (2016)
-
APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages
Nature Communications (2015)