Abstract
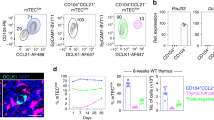
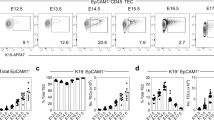
The autoimmune regulator Aire is expressed in a small proportion of medullary thymic epithelial cells (mTECs) and is crucial in the induction of central T cell tolerance. The origin and development of Aire+ mTECs, however, are not well understood. Here we demonstrate that the tight-junction components claudin-3 and claudin-4 (Cld3,4) were 'preferentially' expressed in Aire+ mTECs. In early ontogeny, Cld3,4hi TECs derived from the most apical layer of the stratified thymic anlage first expressed known mTEC markers such as UEA-1 ligand and MTS10. We provide evidence that such Cld3,4hi UEA-1+ TECs represented the initial progenitors specified for Aire+ mTECs, whose development crucially required NF-κB-inducing kinase and the adaptor molecule TRAF6. Our results suggest that Aire+ mTECs represent terminally differentiated cells in a unique lineage arising during thymic organogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
van Ewijk, W. et al. Thymic microenvironments, 3-D versus 2-D? Semin. Immunol. 11, 57–64 (1999).
Gill, J. et al. Thymic generation and regeneration. Immunol. Rev. 195, 28–50 (2003).
Blackburn, C.C. & Manley, N.R. Developing a new paradigm for thymus organogenesis. Nat. Rev. Immunol. 4, 278–289 (2004).
Anderson, G., Owen, J.J., Moore, N.C. & Jenkinson, E.J. Thymic epithelial cells provide unique signals for positive selection of CD4+CD8+ thymocytes in vitro. J. Exp. Med. 179, 2027–2031 (1994).
Petrie, H.T. Cell migration and the control of post-natal T-cell lymphopoiesis in the thymus. Nat. Rev. Immunol. 3, 859–866 (2003).
Takahama, Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 6, 127–135 (2006).
Kyewski, B. & Klein, L. A central role for central tolerance. Annu. Rev. Immunol. 24, 571–606 (2006).
Farr, A.G., Dooley, J.L. & Erickson, M. Organization of thymic medullary epithelial heterogeneity: implications for mechanisms of epithelial differentiation. Immunol. Rev. 189, 20–27 (2002).
Anderson, M.S. et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002).
Derbinski, J. et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202, 33–45 (2005).
Bennett, A.R. et al. Identification and characterization of thymic epithelial progenitor cells. Immunity 16, 803–814 (2002).
Gill, J., Malin, M., Hollander, G.A. & Boyd, R. Generation of a complete thymic microenvironment by MTS24+ thymic epithelial cells. Nat. Immunol. 3, 635–642 (2002).
Rossi, S.W., Jenkinson, W.E., Anderson, G. & Jenkinson, E.J. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature 441, 988–991 (2006).
Gillard, G.O. & Farr, A.G. Contrasting models of promiscuous gene expression by thymic epithelium. J. Exp. Med. 202, 15–19 (2005).
Hollander, G.A. et al. Developmental control point in induction of thymic cortex regulated by a subpopulation of prothymocytes. Nature 373, 350–353 (1995).
van Ewijk, W., Hollander, G., Terhorst, C. & Wang, B. Stepwise development of thymic microenvironments in vivo is regulated by thymocyte subsets. Development 127, 1583–1591 (2000).
Itoi, M., Kawamoto, H., Katsura, Y. & Amagai, T. Two distinct steps of immigration of hematopoietic progenitors into the early thymus anlage. Int. Immunol. 13, 1203–1211 (2001).
Gordon, J. et al. Functional evidence for a single endodermal origin for the thymic epithelium. Nat. Immunol. 5, 546–553 (2004).
Tsukita, S., Furuse, M. & Itoh, M. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2, 285–293 (2001).
Klug, D.B. et al. Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc. Natl. Acad. Sci. USA 95, 11822–11827 (1998).
Sonoda, N. et al. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 147, 195–204 (1999).
Kajiura, F. et al. NF-κB-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J. Immunol. 172, 2067–2075 (2004).
Akiyama, T. et al. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science 308, 248–251 (2005).
Derbinski, J., Schulte, A., Kyewski, B. & Klein, L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2, 1032–1039 (2001).
Kyewski, B. & Derbinski, J. Self-representation in the thymus: an extended view. Nat. Rev. Immunol. 4, 688–698 (2004).
Aaltonen, J. et al. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17, 399–403 (1997).
Nagamine, K. et al. Positional cloning of the APECED gene. Nat. Genet. 17, 393–398 (1997).
Ramsey, C. et al. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum. Mol. Genet. 11, 397–409 (2002).
Kuroda, N. et al. Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J. Immunol. 174, 1862–1870 (2005).
Zhang, M. et al. T cell tolerance to a neo-self antigen expressed by thymic epithelial cells: the soluble form is more effective than the membrane-bound form. J. Immunol. 170, 3954–3962 (2003).
Gallegos, A.M. & Bevan, M.J. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J. Exp. Med. 200, 1039–1049 (2004).
Gillard, G.O. & Farr, A.G. Features of medullary thymic epithelium implicate postnatal development in maintaining epithelial heterogeneity and tissue-restricted antigen expression. J. Immunol. 176, 5815–5824 (2006).
Furuse, M. et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J. Cell Biol. 156, 1099–1111 (2002).
Tsukita, S. & Furuse, M. Claudin-based barrier in simple and stratified cellular sheets. Curr. Opin. Cell Biol. 14, 531–536 (2002).
Rodewald, H.R., Paul, S., Haller, C., Bluethmann, H. & Blum, C. Thymus medulla consisting of epithelial islets each derived from a single progenitor. Nature 414, 763–768 (2001).
Bleul, C.C. et al. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 441, 992–996 (2006).
Chin, R.K. et al. Lymphotoxin pathway directs thymic Aire expression. Nat. Immunol. 4, 1121–1127 (2003).
Boehm, T., Scheu, S., Pfeffer, K. & Bleul, C.C. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTβR. J. Exp. Med. 198, 757–769 (2003).
Zuklys, S. et al. Normal thymic architecture and negative selection are associated with Aire expression, the gene defective in the autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). J. Immunol. 165, 1976–1983 (2000).
Burkly, L. et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 373, 531–536 (1995).
Weih, F. et al. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/Rel family. Cell 80, 331–340 (1995).
Furuse, M., Sasaki, H., Fujimoto, K. & Tsukita, S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 143, 391–401 (1998).
Kobayashi, T. et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity 19, 353–363 (2003).
Ishida, M. et al. Differential expression of PD-L1 and PD-L2, ligands for an inhibitory receptor PD-1, in the cells of lymphohematopoietic tissues. Immunol. Lett. 84, 57–62 (2002).
Niki, S. et al. Alteration of intra-pancreatic target-organ specificity by abrogation of Aire in NOD mice. J. Clin. Invest. 116, 1292–1301 (2006).
Liston, A. et al. Gene dosage–limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J. Exp. Med. 200, 1015–1026 (2004).
Katahira, J., Inoue, N., Horiguchi, Y., Matsuda, M. & Sugimoto, N. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J. Cell Biol. 136, 1239–1247 (1997).
Gray, D.H., Chidgey, A.P. & Boyd, R.L. Analysis of thymic stromal cell populations using flow cytometry. J. Immunol. Methods 260, 15–28 (2002).
Hamazaki, Y., Itoh, M., Sasaki, H., Furuse, M. & Tsukita, S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J. Biol. Chem. 277, 455–461 (2002).
Acknowledgements
We thank S. Tsukita and M. Furuse for providing claudin-transfected L cells; Y. Horiguchi (Research Institute for Microbial Diseases, Osaka University) for the C-CPE plasmid; H. Kawamoto and K. Masuda for discussions; and W.T.V. Germeraad for proofreading the manuscript. Supported by the Ministry of Education, Culture, Science, Sports and Technology of the Japanese government and Shimizu Foundation for Immunology Research.
Author information
Authors and Affiliations
Contributions
Y.H. principally contributed to general experiments and design; H.F. contributed to flow cytometry; T.K. and Y.C. contributed to TRAF6-deficient mice; H.S. contributed to rat monoclonal antibodies to mouse Aire; M.M. contributed to Aire-deficient mice and discussions; and N.M. provided the overall design of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Specific binding of recombinant C-CPE to claudin-3, 4 on the epithelial cell surface. (PDF 341 kb)
Supplementary Fig. 2
Rare Cld3, 4− Aire+ cells express CD11c. (PDF 533 kb)
Supplementary Fig. 3
Phenotypes of the sorted fractions from the B6 thymic stroma cells at E13.5. (PDF 560 kb)
Supplementary Fig. 4
Anti-H-2Kb antibody specifically detects both B6 donor-derived mTECs and cTECs in the reconstituted thymus. (PDF 991 kb)
Supplementary Fig. 5
Transfer of the Cld3,4low TECs at E13.5 results in the generation of all types of TECs including Cld3,4high Aire+ mTECs. (PDF 1473 kb)
Supplementary Table 1
PCR primers. (PDF 20 kb)
Rights and permissions
About this article
Cite this article
Hamazaki, Y., Fujita, H., Kobayashi, T. et al. Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol 8, 304–311 (2007). https://doi.org/10.1038/ni1438
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1438
This article is cited by
-
Human thymoma-associated mutation of the GTF2I transcription factor impairs thymic epithelial progenitor differentiation in mice
Communications Biology (2022)
-
Thymic epithelial cell heterogeneity: TEC by TEC
Nature Reviews Immunology (2020)
-
FoxN1 mediates thymic cortex–medulla differentiation through modifying a developmental pattern based on epithelial tubulogenesis
Histochemistry and Cell Biology (2019)
-
Thymic tolerance as a key brake on autoimmunity
Nature Immunology (2018)
-
Epithelial LTβR signaling controls the population size of the progenitors of medullary thymic epithelial cells in neonatal mice
Scientific Reports (2017)