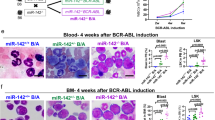
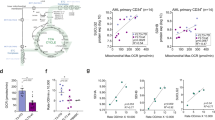
Abstract
Expression of the MECOM (also known as EVI1) proto-oncogene is deregulated by chromosomal translocations in some cases of acute myeloid leukemia (AML) and is associated with poor clinical outcome. Here, through transcriptomic and metabolomic profiling of hematopoietic cells, we reveal that EVI1 overexpression alters cellular metabolism. A screen using pooled short hairpin RNAs (shRNAs) identified the ATP-buffering, mitochondrial creatine kinase CKMT1 as necessary for survival of EVI1-expressing cells in subjects with EVI1-positive AML. EVI1 promotes CKMT1 expression by repressing the myeloid differentiation regulator RUNX1. Suppression of arginine–creatine metabolism by CKMT1-directed shRNAs or by the small molecule cyclocreatine selectively decreased the viability, promoted the cell cycle arrest and apoptosis of human EVI1-positive cell lines, and prolonged survival in both orthotopic xenograft models and mouse models of primary AML. CKMT1 inhibition altered mitochondrial respiration and ATP production, an effect that was abrogated by phosphocreatine-mediated reactivation of the arginine–creatine pathway. Targeting CKMT1 is thus a promising therapeutic strategy for this EVI1-driven AML subtype that is highly resistant to current treatment regimens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Baysal, B.E. et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851 (2000).
Janeway, K.A. et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc. Natl. Acad. Sci. USA 108, 314–318 (2011).
Ricketts, C. et al. Germline SDHB mutations and familial renal cell carcinoma. J. Natl. Cancer Inst. 100, 1260–1262 (2008).
Kim, S., Kim, D.H., Jung, W.H. & Koo, J.S. Succinate dehydrogenase expression in breast cancer. Springerplus 2, 299 (2013).
Cairns, R.A. & Mak, T.W. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models and clinical opportunities. Cancer Discov. 3, 730–741 (2013).
Figueroa, M.E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
Lu, C. et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 (2012).
Ward, P.S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Cantor, J.R. & Sabatini, D.M. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2, 881–898 (2012).
Burnett, A., Wetzler, M. & Löwenberg, B. Therapeutic advances in acute myeloid leukemia. J. Clin. Oncol. 29, 487–494 (2011).
Patel, J.P. et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366, 1079–1089 (2012).
Glass, C., Wilson, M., Gonzalez, R., Zhang, Y. & Perkins, A.S. The role of EVI1 in myeloid malignancies. Blood Cells Mol. Dis. 53, 67–76 (2014).
Goyama, S. & Kurokawa, M. Evi1 as a critical regulator of leukemic cells. Int. J. Hematol. 91, 753–757 (2010).
Gröschel, S. et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J. Clin. Oncol. 28, 2101–2107 (2010).
Lugthart, S. et al. Clinical, molecular and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J. Clin. Oncol. 28, 3890–3898 (2010).
Kustikova, O.S. et al. Activation of Evi1 inhibits cell cycle progression and differentiation of hematopoietic progenitor cells. Leukemia 27, 1127–1138 (2013).
Goyama, S. et al. Evi1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell 3, 207–220 (2008).
Kataoka, K. et al. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J. Exp. Med. 208, 2403–2416 (2011).
Bard-Chapeau, E.A. et al. Ecotopic viral integration site 1 (EVI1) regulates multiple cellular processes important for cancer and is a synergistic partner for FOS protein in invasive tumors. Proc. Natl. Acad. Sci. USA 109, 2168–2173 (2012).
Glass, C. et al. Global identification of EVI1 target genes in acute myeloid leukemia. PLoS One 8, e67134 (2013).
Furter, R., Furter-Graves, E.M. & Wallimann, T. Creatine kinase: the reactive cysteine is required for synergism but is non-essential for catalysis. Biochemistry 32, 7022–7029 (1993).
Burgess, M.R. et al. Preclinical efficacy of MEK inhibition in Nras-mutant AML. Blood 124, 3947–3955 (2014).
Li, Q. et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 117, 2022–2032 (2011).
Herranz, D. et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat. Med. 21, 1182–1189 (2015).
Wang, Y.H. et al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 158, 1309–1323 (2014).
Mussai, F. et al. Arginine dependence of acute myeloid leukemia blast proliferation: a novel therapeutic target. Blood 125, 2386–2396 (2015).
Plunkett, W. Arginine addiction in AML. Blood 125, 3971–3972 (2015).
Miraki-Moud, F. et al. Arginine deprivation using pegylated arginine deiminase has activity against primary acute myeloid leukemia cells in vivo. Blood 125, 4060–4068 (2015).
Wallimann, T., Wyss, M., Brdiczka, D., Nicolay, K. & Eppenberger, H.M. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the 'phosphocreatine circuit' for cellular energy homeostasis. Biochem. J. 281, 21–40 (1992).
Roberts, D.J. & Miyamoto, S. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 22, 248–257 (2015).
Arora, K.K. & Pedersen, P.L. Functional significance of mitochondrial-bound hexokinase in tumor cell metabolism. Evidence for preferential phosphorylation of glucose by intramitochondrially generated ATP. J. Biol. Chem. 263, 17422–17428 (1988).
Mathupala, S.P., Ko, Y.H. & Pedersen, P.L. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene 25, 4777–4786 (2006).
Wyss, M. & Kaddurah-Daouk, R. Creatine and creatinine metabolism. Physiol. Rev. 80, 1107–1213 (2000).
Cimino, D. et al. Identification of new genes associated with breast cancer progression by gene expression analysis of predefined sets of neoplastic tissues. Int. J. Cancer 123, 1327–1338 (2008).
Gaidzik, V.I. et al. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J. Clin. Oncol. 29, 1364–1372 (2011).
Zuber, J. et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 29, 79–83 (2011).
Banerji, V. et al. The intersection of genetic and chemical genomic screens identifies GSK-3α as a target in human acute myeloid leukemia. J. Clin. Invest. 122, 935–947 (2012).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Luo, B. et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. USA 105, 20380–20385 (2008).
Ben-Sahra, I., Howell, J.J., Asara, J.M. & Manning, B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328 (2013).
Jones, S.J. Prediction of genomic functional elements. Annu. Rev. Genomics Hum. Genet. 7, 315–338 (2006).
Thorvaldsdóttir, H., Robinson, J.T. & Mesirov, J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
Bernt, K.M. et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011).
Puissant, A. et al. SYK is a critical regulator of FLT3 in acute myeloid leukemia. Cancer Cell 25, 226–242 (2014).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Mootha, V.K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Merico, D., Isserlin, R. & Bader, G.D. Visualizing gene-set-enrichment results using the Cytoscape plug-in enrichment map. Methods Mol. Biol. 781, 257–277 (2011).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G.D. Enrichment map: a network-based method for gene-set-enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Leng, N. et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043 (2013).
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011).
Stegmaier, K. et al. Gene-expression-based high-throughput screening (GE–HTS) and application to leukemia differentiation. Nat. Genet. 36, 257–263 (2004).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukemia. Nature 478, 524–528 (2011).
Daigle, S.R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011).
Wouters, B.J. et al. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 113, 3088–3091 (2009).
Tomasson, M.H. et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 111, 4797–4808 (2008).
Acknowledgements
We thank T. Sato and M. Kurokawa (University of Tokyo) for providing plasmid constructs and detailed procedures for Evi1 overexpression in mouse hematopoietic cells. We also thank J.F. Clark (University of Cincinnati) for advice on the use of cyclocreatine in vivo. This research was supported with grants from the US National Cancer Institute (NCI) (NIH 1R35 CA210030-01 (K. Stegmaier) and R37 CA72614 (K. Shannon)), the Stand-up-to-Cancer Program (K. Stegmaier); the Bridge Project, a collaboration between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber–Harvard Cancer Center (DF–HCC) (K. Stegmaier and M.T.H.) and the Koch Institute Cancer Center Support (NCI grant P30-CA14051; M.T.H.), and with support from the Cubans Curing Children's Cancers (4C's Fund) (K. Stegmaier). A.P. is a recipient of support from the ATIP–AVENIR and LNCC French research programs, the EHA research grant for a Non-Clinical Advanced fellow, and is supported by the St. Louis Association for leukemia research. K. Stegmaier is an LLS Scholar. A.P., N.F. and I.B.-S. were awarded the 'Prix Jeune Chercheur' from the Bettencourt Foundation and the Franco-American Exchange Prize from Philippe Foundation Inc.
Author information
Authors and Affiliations
Contributions
N.F. and C.F.B. contributed equally to the manuscript as joint first authors. N.F. and C.F.B. developed the study, established conditions for in vivo and in vitro experiments, acquired and analyzed the data, and wrote the manuscript; I.B.-S. designed and performed the metabolism-related experiments; L.B., A.R., Y.P., A.S.C. and F.L. designed, performed, and analyzed the in vivo experiments; Q.L., M.R.B. and K. Shannon revised the manuscript and provided NrasG12D and NrasG12D + Evi1 mouse models and methodology for in vivo functional analyses; G.A. revised the manuscript and performed statistical analysis, biostatistics, and computational analysis of the RNA sequencing, the publicly available patient sample cohorts, and the shRNA screening experiments; A.S.P. and Y.Z. provided reagents and ChIP-seq data for the ChIP–qPCR experiments performed on endogenous mouse Evi1; I.G., D.J.D. and R.M.S. provided patient samples and revised the manuscript; P.A. revised the manuscript; M.T.H., A.P. and K Stegmaier contributed equally to this work as joint senior authors; M.T.H., A.P. and K. Stegmaier supervised the study, wrote and revised the manuscript, designed the in vitro and in vivo experiments, analyzed the data and provided funding for the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–8 and Supplementary Note (PDF 19995 kb)
Rights and permissions
About this article
Cite this article
Fenouille, N., Bassil, C., Ben-Sahra, I. et al. The creatine kinase pathway is a metabolic vulnerability in EVI1-positive acute myeloid leukemia. Nat Med 23, 301–313 (2017). https://doi.org/10.1038/nm.4283
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4283
This article is cited by
-
Mitochondrial creatine kinase 1 regulates the cell cycle in non-small cell lung cancer via activation of cyclin-dependent kinase 4
Respiratory Research (2023)
-
A creatine kinase inhibitor targeting a redox-regulated cysteine
Nature Chemical Biology (2023)
-
Depletion of creatine phosphagen energetics with a covalent creatine kinase inhibitor
Nature Chemical Biology (2023)
-
Reduced-intensity conditioning is effective for allogeneic hematopoietic stem cell transplantation in infants with MECOM-associated syndrome
International Journal of Hematology (2023)
-
Acidic ascites inhibits ovarian cancer cell proliferation and correlates with the metabolomic, lipidomic and inflammatory phenotype of human patients
Journal of Translational Medicine (2022)