Abstract
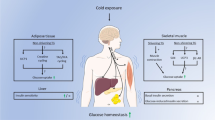
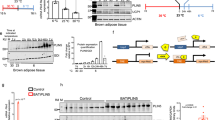
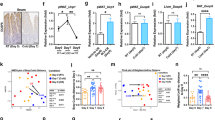
Adaptive thermogenesis is an energy-demanding process that is mediated by cold-activated beige and brown adipocytes, and it entails increased uptake of carbohydrates, as well as lipoprotein-derived triglycerides and cholesterol, into these thermogenic cells. Here we report that cold exposure in mice triggers a metabolic program that orchestrates lipoprotein processing in brown adipose tissue (BAT) and hepatic conversion of cholesterol to bile acids via the alternative synthesis pathway. This process is dependent on hepatic induction of cytochrome P450, family 7, subfamily b, polypeptide 1 (CYP7B1) and results in increased plasma levels, as well as fecal excretion, of bile acids that is accompanied by distinct changes in gut microbiota and increased heat production. Genetic and pharmacological interventions that targeted the synthesis and biliary excretion of bile acids prevented the rise in fecal bile acid excretion, changed the bacterial composition of the gut and modulated thermogenic responses. These results identify bile acids as important metabolic effectors under conditions of sustained BAT activation and highlight the relevance of cholesterol metabolism by the host for diet-induced changes of the gut microbiota and energy metabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Turnbaugh, P.J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006).
Musso, G., Gambino, R. & Cassader, M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 62, 361–380 (2011).
Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185 (2012).
Ridaura, V.K. et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013).
Wang, Z. et al. Nonlethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015).
Zhu, W. et al. Gut microbial metabolite TMAO enhances platelet hyper-reactivity and thrombosis risk. Cell 165, 111–124 (2016).
Schwabe, R.F. & Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 13, 800–812 (2013).
David, L.A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Carmody, R.N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Chevalier, C. et al. Gut microbiota orchestrates energy homeostasis during cold. Cell 163, 1360–1374 (2015).
Ziętak, M. et al. Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab. 23, 1216–1223 (2016).
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Harms, M. & Seale, P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013).
Bartelt, A. & Heeren, J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 10, 24–36 (2014).
Bartelt, A. et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205 (2011).
Stanford, K.I. et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J. Clin. Invest. 123, 215–223 (2013).
Berbée, J.F. et al. Brown fat activation reduces hypercholesterolemia and protects from atherosclerosis development. Nat. Commun. 6, 6356 (2015).
Russell, D.W. The enzymes, regulation and genetics of bile acid synthesis. Annu. Rev. Biochem. 72, 137–174 (2003).
Li-Hawkins, J., Lund, E.G., Turley, S.D. & Russell, D.W. Disruption of the oxysterol 7α-hydroxylase gene in mice. J. Biol. Chem. 275, 16536–16542 (2000).
Kuipers, F., Bloks, V.W. & Groen, A.K. Beyond intestinal soap—bile acids in metabolic control. Nat. Rev. Endocrinol. 10, 488–498 (2014).
Kalaany, N.Y. & Mangelsdorf, D.J. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 68, 159–191 (2006).
Pols, T.W., Noriega, L.G., Nomura, M., Auwerx, J. & Schoonjans, K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol. 54, 1263–1272 (2011).
Katsuma, S., Hirasawa, A. & Tsujimoto, G. Bile acids promote glucagon-like peptide 1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem. Biophys. Res. Commun. 329, 386–390 (2005).
Watanabe, M. et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489 (2006).
Broeders, E.P. et al. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab. 22, 418–426 (2015).
Joyce, S.A. et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 111, 7421–7426 (2014).
Islam, K.B. et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141, 1773–1781 (2011).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108 (2012).
Sonnenburg, E.D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Hambruch, E. et al. Synthetic farnesoid X receptor agonists induce high-density-lipoprotein-mediated transhepatic cholesterol efflux in mice and monkeys and prevent atherosclerosis in cholesteryl ester transfer protein transgenic low-density lipoprotein receptor−/− mice. J. Pharmacol. Exp. Ther. 343, 556–567 (2012).
Nedergaard, J., Bengtsson, T. & Cannon, B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293, E444–E452 (2007).
Cypess, A.M. et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 (2009).
Saito, M. et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531 (2009).
van Marken Lichtenbelt, W.D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009).
Ouellet, V. et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Invest. 122, 545–552 (2012).
Nedergaard, J. & Cannon, B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 11, 268–272 (2010).
Schlein, C. et al. FGF21 lowers plasma triglycerides by accelerating lipoprotein catabolism in white and brown adipose tissues. Cell Metab. 23, 441–453 (2016).
Cannon, C.P. et al. Ezetimibe added to statin therapy after acute coronary syndromes. N. Engl. J. Med. 372, 2387–2397 (2015).
Levy, M. et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443 (2015).
Wang, G.X. et al. The brown-fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat. Med. 20, 1436–1443 (2014).
Scheja, L. & Heeren, J. Metabolic interplay between white, beige, brown adipocytes and the liver. J. Hepatol. 64, 1176–1186 (2016).
Sayin, S.I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-β-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Zhong, C.Y. et al. Microbiota prevents cholesterol loss from the body by regulating host gene expression in mice. Sci. Rep. 5, 10512 (2015).
Brufau, G. et al. Improved glycemic control with colesevelam treatment in patients with type 2 diabetes is not directly associated with changes in bile acid metabolism. Hepatology 52, 1455–1464 (2010).
Haeusler, R.A., Astiarraga, B., Camastra, S., Accili, D. & Ferrannini, E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes 62, 4184–4191 (2013).
Hanssen, M.J. et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat. Med. 21, 863–865 (2015).
Rohlmann, A., Gotthardt, M., Hammer, R.E. & Herz, J. Inducible inactivation of hepatic LRP gene by Cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants. J. Clin. Invest. 101, 689–695 (1998).
Rühlemann, M.C. et al. Fecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut 66, 753–754 (2017).
Wang, Y., Naumann, U., Wright, S.T. & Warton, D.I. mvabund—an R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 3, 471–474 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
John, C. et al. A liquid chromatography–tandem mass spectrometry–based method for the simultaneous determination of hydroxy sterols and bile acids. J. Chromatogr. A 1371, 184–195 (2014).
Eissing, L. et al. De novo lipogenesis in human fat and liver is linked to ChREBP-β and metabolic health. Nat. Commun. 4, 1528 (2013).
Meiss, E. et al. Metabolite targeting: development of a comprehensive targeted metabolomics platform for the assessment of diabetes and its complications. Metabolomics 12, 52 (2016).
Acknowledgements
We thank S. Ehret, B. Henkel, A. Kuhl and E.-M. Azizi for excellent technical assistance, P. Dawson (Emory University School of Medicine) for the ASBT-specific polyclonal antibody, and J. Nedergaard and B. Cannon (Wenner-Gren Institute, Stockholm University) for the UCP1-spcific polyclonal antibody. This work was supported by grants funded by the Deutsche Forschungsgemeinschaft (SFB841, “Liver inflammation: infection, immune regulation und consequences” (J.H. and M.D.); KFO306, “Primary sclerosing cholangitis (J.H. and to A.F.)), a Heisenberg Professorship (HE3645/7-1 (J.H.) and DA1063/3-2 (M.D.)), an EFSD award supported by Merck Sharp Dohme (MSD) (J.H.), the EU FP7 project RESOLVE FP7-HEALTH-2012-305707 (J.H.), a University Medical Center Hamburg–Eppendorf MD/PhD fellowship (C.S.) and the US National Institutes of Health grant HL087564 (P.W.S.).
Author information
Authors and Affiliations
Contributions
A.W., C.J., L.S. and J.H. designed the study, were involved in all aspects of the experiments and wrote the manuscript; M.C.R., F.-A.H. and A.F. were responsible for the microbiome analysis; M.B., N.S., M.H., I.E., C.S. and C.M. were involved in the metabolic studies; M.F., M.D., A.F., C.K. und P.W.S. were involved in study design; and all authors read and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
C.K. is a co-founder and shareholder of Phenex Pharmaceuticals AG.
Supplementary information
Supplementary Figures and Table
Supplementary Figures 1–15 and Supplementary Table 1 (PDF 2708 kb)
Rights and permissions
About this article
Cite this article
Worthmann, A., John, C., Rühlemann, M. et al. Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nat Med 23, 839–849 (2017). https://doi.org/10.1038/nm.4357
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4357
This article is cited by
-
Intestinal dual-specificity phosphatase 6 regulates the cold-induced gut microbiota remodeling to promote white adipose browning
npj Biofilms and Microbiomes (2024)
-
Caloric restriction remodels the hepatic chromatin landscape and bile acid metabolism by modulating the gut microbiota
Genome Biology (2023)
-
Discovery biomarker to optimize obeticholic acid treatment for non-alcoholic fatty liver disease
Biology Direct (2023)
-
Characteristics of glucose and lipid metabolism and the interaction between gut microbiota and colonic mucosal immunity in pigs during cold exposure
Journal of Animal Science and Biotechnology (2023)
-
Gut microbiome signatures of extreme environment adaption in Tibetan pig
npj Biofilms and Microbiomes (2023)