Abstract
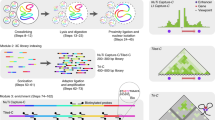
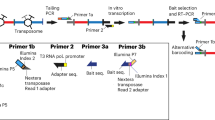
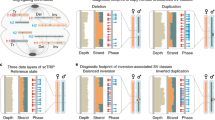
Balanced chromosomal rearrangements can cause disease, but techniques for their rapid and accurate identification are missing. Here we demonstrate that chromatin conformation capture on chip (4C) technology can be used to screen large genomic regions for balanced and complex inversions and translocations at high resolution. The 4C technique can be used to detect breakpoints also in repetitive DNA sequences as it uniquely relies on capturing genomic fragments across the breakpoint. Using 4C, we uncovered LMO3 as a potentially leukemogenic translocation partner of TRB@. We developed multiplex 4C to simultaneously screen for translocation partners of multiple selected loci. We identified unsuspected translocations and complex rearrangements. Furthermore, using 4C we detected translocations even in small subpopulations of cells. This strategy opens avenues for the rapid fine-mapping of cytogenetically identified translocations and inversions, and the efficient screening for balanced rearrangements near candidate loci, even when rearrangements exist only in subpopulations of cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Iafrate, A.J. et al. Detection of large-scale variation in the human genome. Nat. Genet. 36, 949–951 (2004).
Mehan, M.R., Freimer, N.B. & Ophoff, R.A. A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum. Genomics 1, 335–344 (2004).
Sharp, A.J., Cheng, Z. & Eichler, E.E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 7, 407–442 (2006).
Easton, D.F. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093 (2007).
Eichler, E.E. et al. Completing the map of human genetic variation. Nature 447, 161–165 (2007).
Feuk, L., Carson, A.R. & Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97 (2006).
Campbell, P.J. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40, 722–729 (2008).
Korbel, J.O. et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 318, 420–426 (2007).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Simonis, M., Kooren, J. & de Laat, W. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods 4, 895–901 (2007).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Burnett, R.C., Thirman, M.J., Rowley, J.D. & Diaz, M.O. Molecular analysis of the T-cell acute lymphoblastic leukemia-associated t(1;7)(p34;q34) that fuses LCK and TCRB. Blood 84, 1232–1236 (1994).
Soulier, J. et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood 106, 274–286 (2005).
Speleman, F. et al. A new recurrent inversion, inv(7)(p15q34), leads to transcriptional activation of HOXA10 and HOXA11 in a subset of T-cell acute lymphoblastic leukemias. Leukemia 19, 358–366 (2005).
Galjaard, R.J. et al. Isolated postaxial polydactyly type B with mosaicism of a submicroscopic unbalanced translocation leading to an extended phenotype in offspring. Am. J. Med. Genet. A. 121, 168–173 (2003).
Aoyama, M. et al. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 65, 4587–4597 (2005).
Armstrong, S.A. & Look, A.T. Molecular genetics of acute lymphoblastic leukemia. J. Clin. Oncol. 23, 6306–6315 (2005).
Graux, C., Cools, J., Michaux, L., Vandenberghe, P. & Hagemeijer, A. Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: from thymocyte to lymphoblast. Leukemia 20, 1496–1510 (2006).
Hatano, M., Roberts, C.W. & Minden, M. Crist. W.M. and Korsmeyer, S.J. Deregulation of a homeobox gene, HOX11, by the t(10;14) in T cell leukemia. Science 253, 79–82 (1991).
Kennedy, M.A. et al. HOX11, a homeobox-containing T-cell oncogene on human chromosome 10q24. Proc. Natl. Acad. Sci. USA 88, 8900–8904 (1991).
Boehm, T. Foroni, L., Kaneko, Y., Perutz, M.F. & Rabbitts, T.H. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc. Natl. Acad. Sci. USA 88, 4367–4371 (1991).
Van Vlierberghe, P. et al. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood 108, 3520–3529 (2006).
Ling, J.Q. et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312, 269–272 (2006).
Lomvardas, S. et al. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 (2006).
Wurtele, H. & Chartrand, P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 14, 477–495 (2006).
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Tomlins, S.A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005).
Chen, W. et al. Mapping translocation breakpoints by next-generation sequencing. Genome Res. 18, 1143–1149 (2008).
Fiegler, H. et al. Array painting: a method for the rapid analysis of aberrant chromosomes using DNA microarrays. J. Med. Genet. 40, 664–670 (2003).
van Vlierberghe, P. et al. A new recurrent 9q34 duplication in pediatric T-cell acute lymphoblastic leukemia. Leukemia 20, 1245–1253 (2006).
Acknowledgements
We thank A. de Klein, W. van Ijcken, J. Gladdines, J. Veltman, A. Hoischen and E. Splinter for assistance. This work was supported by grants from the Dutch Cancer Society (KWF-EMCR 2006-3500) to J.P.P.M., grants from the European Union and the Cancer Genomics Centre to F.G., grants from the Dutch Scientific Organization (NWO) (912-04-082), Netherlands Genomics Initiative (050-71-324) and the Erasmus MC to W.d.L.
Author information
Authors and Affiliations
Contributions
M.S. designed, performed and analyzed experiments and wrote the manuscript; P.K. performed and analyzed experiments; I.H. helped perform experiments; R.-J.G.: provided PAP cell line and helped design the PAP experiment; E.-J.R. designed the microarray. F.G. helped design experiments; J.P.P.M. provided leukemia samples, helped design T-ALL experiments and helped write the manuscript; and W.d.L. designed experiments, supervised the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
W.d.L. and F.G hold patent applications PCT/IB2006/002268 and PCT/IB2008/000625 on 4C technology (published by World Intellectual Property Organization).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Table 1 (PDF 6740 kb)
Rights and permissions
About this article
Cite this article
Simonis, M., Klous, P., Homminga, I. et al. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nat Methods 6, 837–842 (2009). https://doi.org/10.1038/nmeth.1391
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1391
This article is cited by
-
Robust detection of translocations in lymphoma FFPE samples using targeted locus capture-based sequencing
Nature Communications (2021)
-
Discovery of regulatory noncoding variants in individual cancer genomes by using cis-X
Nature Genetics (2020)
-
3C and 3C-based techniques: the powerful tools for spatial genome organization deciphering
Molecular Cytogenetics (2018)
-
Chromatin conformation analysis of primary patient tissue using a low input Hi-C method
Nature Communications (2018)
-
Three-dimensional genome architecture and emerging technologies: looping in disease
Genome Medicine (2017)