Abstract
We developed molecular tension probes (TPs) that report traction forces of adherent cells with high spatial resolution, can in principle be linked to virtually any surface, and obviate monitoring deformations of elastic substrates. TPs consist of DNA hairpins conjugated to fluorophore-quencher pairs that unfold and fluoresce when subjected to specific forces. We applied TPs to reveal that cellular traction forces are heterogeneous within focal adhesions and localized at their distal edges.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Beningo, K.A. & Wang, Y.L. Trends Cell Biol. 12, 79–84 (2002).
Mammoto, T. & Ingber, D.E. Development 137, 1407–1420 (2010).
McBeath, R., Pirone, D.M., Nelson, C.M., Bhadriraju, K. & Chen, C.S. Dev. Cell 6, 483–495 (2004).
Pirone, D.M. et al. J. Cell Biol. 174, 277–288 (2006).
Dembo, M. & Wang, Y.L. Biophys. J. 76, 2307–2316 (1999).
Tan, J.L. et al. Proc. Natl. Acad. Sci. USA 100, 1484–1489 (2003).
Engler, A.J., Sen, S., Sweeney, H.L. & Discher, D.E. Cell 126, 677–689 (2006).
Paszek, M.J. et al. Cancer Cell 8, 241–254 (2005).
Pelham, R.J. Jr. & Wang, Y. Proc. Natl. Acad. Sci. USA 94, 13661–13665 (1997).
Legant, W.R. et al. Nat. Methods 7, 969–971 (2010).
Liu, Y., Yehl, K., Narui, Y. & Salaita, K. J. Am. Chem. Soc. 135, 5320–5323 (2013).
Wang, X. & Ha, T. Science 340, 991–994 (2013).
Kong, F., García, A.J., Mould, A.P., Humphries, M.J. & Zhu, C. J. Cell Biol. 185, 1275–1284 (2009).
Sun, Z. et al. Am. J. Physiol. Heart Circ. Physiol. 289, H2526–H2535 (2005).
Woodside, M.T. et al. Science 314, 1001–1004 (2006).
Woodside, M.T. et al. Proc. Natl. Acad. Sci. USA 103, 6190–6195 (2006).
Ruoslahti, E. & Pierschbacher, M.D. Cell 44, 517–518 (1986).
Anthony, P.C. et al. J. Am. Chem. Soc. 134, 4607–4614 (2012).
Balaban, N.Q. et al. Nat. Cell Biol. 3, 466–472 (2001).
Uehata, M. et al. Nature 389, 990–994 (1997).
Yoshioka, K., Matsumura, F., Akedo, H. & Itoh, K. J. Biol. Chem. 273, 5146–5154 (1998).
Reinhart-King, C.A., Dembo, M. & Hammer, D.A. Langmuir 19, 1573–1579 (2003).
Sabass, B., Gardel, M.L., Waterman, C.M. & Schwarz, U.S. Biophys. J. 94, 207–220 (2008).
Plotnikov, S.V., Pasapera, A.M., Sabass, B. & Waterman, C.M. Cell 151, 1513–1527 (2012).
Legant, W.R. et al. Proc. Natl. Acad. Sci. USA 110, 881–886 (2013).
Style, R.W. et al. Soft Matter 10, 4047–4055 (2014).
Grashoff, C. et al. Nature 466, 263–266 (2010).
Stabley, D.R., Jurchenko, C., Marshall, S.S. & Salaita, K.S. Nat. Methods 9, 64–67 (2012).
Zimmermann, J.L., Nicolaus, T., Neuert, G. & Blank, K. Nat. Protoc. 5, 975–985 (2010).
Sigal, G.B., Mrksich, M. & Whitesides, G.M. J. Am. Chem. Soc. 120, 3464–3473 (1998).
Acknowledgements
This work was supported in part from grants from the US National Institutes of Health (EB00262, EB08396, EB001046, HL115553, GM065865, GM74048 and GM57035) and Howard Hughes Medical Institute. We thank M. Woodside for assistance in modeling hairpin opening forces. We thank R. Assoian (University of Pennsylvania) for providing MEFs. B.L.B., C.K.C., B.M.B. and C.E.D. acknowledge fellowships from the US National Science Foundation, American Heart Association, Ruth L. Kirschstein National Research Service Award and Novartis Foundation, respectively.
Author information
Authors and Affiliations
Contributions
B.L.B., C.E.D., C.S.C. and D.R.L. conceived of and initiated the project. B.L.B., C.E.D., B.T., L.M.M., C.K.C, P.C.A., V.K.D. and B.M.B. designed, performed, analyzed and interpreted experiments. C.E.D., B.T. and L.M.M. synthesized TPs. P.C.A. and V.K.D. characterized TP mechanics. B.L.B., B.T., C.K.C. and B.M.B. performed the cell-based experiments. S.M.B., C.S.C. and D.R.L. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Chemical synthesis and characterization of TPs.
(a) Detailed chemical scheme for synthesis of TPs. TPs were synthesized using solid-phase synthesis in two fragments. The 3′ end contained a free thiol and the 5′ end of the other fragment was conjugated to the GGRGDS peptide. After purification, the fragments were ligated. (b) Detailed chemical scheme for surface conjugation. The substrate was functionalized with an amine-presenting silane that was coupled to a succinimide-PEG-maleimide crosslinker that was then reacted with the 3′ end of the hairpins. (c) LC/MS analysis of synthesis product. Peaks are located near the calculated expected mass of the TPs. (d) Plot of fluorescence of TPs with temperature. Thermally melting the TPs demonstrated their unfolding-induced fluorescence. Urea was used for sequences with higher melting points.
Supplementary Figure 2 Surface functionalization with TPs is homogeneous.
Raw 60X TIRF fluorescence image of glass functionalized with unstructured linear DNA coupled with fluorescein to demonstrate even coating on the surface.
Supplementary Figure 3 Specific RGD-integrin interaction is required for traction force-induced TP fluorescence.
A negative control TP was synthesized lacking the RGD peptide. Surfaces coated with just this TP failed to support cell adhesion. To allow for cell adhesion to substrate in the presence of this TP, RGD was coupled to the surface through a PEG crosslinker and mixed with equal molar amounts of the negative control TP. Cells spread readily on these surfaces, but no cell-induced fluorescence was observed. Together, these results confirmed that the only cell adhesive interaction with the TP is through RGD.
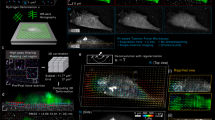
Supplementary Figure 4 Image processing.
Top row shows work-flow for processing raw images sequentially by removing background signal, Wiener filtering, and then band-pass filtering for adhesions. Magenta line shows cross-section used to display how signal intensity was altered at each step (bottom graph). Background from unfolded hairpin was not significantly above bare glass (approximately 4000 on a 16-bit scale), and the signal from FA opening of hairpins reached values 10x above background (approximately 50,000 on a 16-bit scale) for our usual imaging conditions (CFI Apo TIRF 60X oil (1.49NA) objective, 500 msec exposures, and a laser intensity of approximately 190 µW at 488 nm wavelength).
Supplementary Figure 5 TPs conjugated to different dyes are equally functional.
We have developed three TP-dyes. In addition to the fluorescein TPs shown in the main text, we have synthesized fully functional TPs labeled with Alexa 546 demonstrated in (a) and Alexa 647 demonstrated in (b).
Supplementary Figure 6 Measurement of traction forces using two sequences.
(a) Schematic (left) and image (right) of surfaces conjugated with equimolar mixtures of two TPs: one sequence with an F1/2 value of ~9 pN (TP9) labeled with Alexa 647 and another with ~17 pN (TP17) labeled with fluorescein. Images show co-localized adhesion-like traction stress signals in two separate fluorescent channels. The two channels reported different intensity values that result in part from differences in fluorophore and wavelength-dependent optics. (b) To control for these differences, signals were obtained for substrates conjugated with equimolar mixtures of TP9-fluorescein and TP9-Alexa 647. This setup also yielded adhesion-like traction stress signals in two separate fluorescent channels. (c-d) Plot of the intensity of the two colors for each pixel for TP9 Alexa 647 versus TP9 fluorescein (c) and TP9 Alexa 647 versus TP17 fluorescein (d). n ≥ 6 cells, 60 adhesions for each condition. These pixel values changed linearly with the unfolding force of TPs, showing no saturation of signal for the range of cellular forces observed (fitted lines from Theil-Senn estimator).
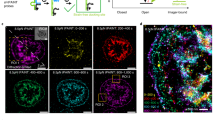
Supplementary Figure 7 Localization of TP-reported stress with different focal adhesion proteins.
Fluorescence of both TPs and transfected cells expressing (a) mRFP-vinculin or (b) mApple-paxillin. Images show the localization of TP-measured tractions to focal adhesions in both cases. The use of different sequences with different F1/2 values did not change this localization.
Supplementary Figure 8 Cell spread area as a function of TP density.
Spread area of cells cultured on surfaces coupled with TP8 at 3 µM, 1 µM, and 0.25 µM. Red lines in box plot mark the median, and whiskers show the ±2.7σ range. n ≥ 8, 185 adhesions for each condition.
Supplementary Figure 9 TP reported forces with diminished surface concentrations of RGD peptide.
Traction forces reported for cells spread on surfaces conjugated in (a) 3 µM, (b) 1 µM, and (c) 0.25 µM of TP. With decreasing force probe concentration, the total force measured for cells (d) and number of adhesions (e) decreases accordingly. (f) Individual adhesion size did not differ significantly between 3 µM and 1 µM TP concentration, but was larger than in cells cultured on 0.25 µM TP6 surfaces. (g) The force per adhesion was highest for cells cultured on 1 µM concentrations of TP6 and lowest for 0.25 µM. (h) Histogram of pixel frequency versus force for each concentration shows shift to higher forces at 1 µM concentration (p-value reported from Wilcoxon rank sum test). Red lines in box plot mark the median, and whiskers show the ±2.7σ range. n ≥ 8, 185 adhesions for each condition.
Supplementary Figure 10 Application of TPs on substrates with complex topography.
DIC (a) and thresholded fluorescence (b) images of a polarized fibroblast on substrates composed of raised 2 um wide PDMS ridges stamped with TP6. Cell is outlined in yellow and ridges are false-colored magenta. Scale bar is 10 µm. (c) Angular histogram of the distribution of force-bearing adhesions as reported by HPs with respect to the axis of polarization. The mean angle of n=52 adhesions analyzed was 25º (red dashed line).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–5 (PDF 2478 kb)
TP-reported traction force dynamics under a cell.
Time-lapse video showing traction dynamics of a cell attached to the substrate. Left panel shows the fluorescence images captured in TIRF mode using a 60x objective. The following three panels show the stress maps obtained from these images. Scale bar is 20 μm in the top two panels and 8 μm for the bottom two panels. The length of the video is 62 min. (MOV 6950 kb)
TP-reported traction force dynamics following inhibition of cellular contractility.
Time-lapse video showing traction dynamics of a cell attached to the substrate that was then treated with ROCK inhibitor Y27632. Left panel shows the fluorescence images captured in TIRF mode using a 60x objective. The following two panels show the stress maps obtained from these images. Scale bar is 20 μm for the left two panels and 8 μm for the right panel. The recording begins immediately following introduction of Y27632. The length of the video is 26 min. (MOV 2192 kb)
TP-reported traction force dynamics following stimulation of cellular contractility.
Time-lapse video showing traction dynamics of a cell attached to the substrate that was then treated with LPA. Left panel shows the fluorescence images captured in TIRF mode using a 60x objective. The following two panels show the stress maps obtained from these images. The recording begins immediately following introduction of LPA. Scale bar is 20 μm for the left two panels and 8 μm for the right panel. The length of the video is 60 min. (MOV 1281 kb)
Rights and permissions
About this article
Cite this article
Blakely, B., Dumelin, C., Trappmann, B. et al. A DNA-based molecular probe for optically reporting cellular traction forces. Nat Methods 11, 1229–1232 (2014). https://doi.org/10.1038/nmeth.3145
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3145
This article is cited by
-
RAD-TGTs: high-throughput measurement of cellular mechanotype via rupture and delivery of DNA tension probes
Nature Communications (2023)
-
Hydrogel-based molecular tension fluorescence microscopy for investigating receptor-mediated rigidity sensing
Nature Methods (2023)
-
Single-molecule characterization of subtype-specific β1 integrin mechanics
Nature Communications (2022)
-
A reversible shearing DNA probe for visualizing mechanically strong receptors in living cells
Nature Cell Biology (2021)
-
Turn-key mapping of cell receptor force orientation and magnitude using a commercial structured illumination microscope
Nature Communications (2021)