Abstract
The Q system is a repressible binary expression system for transgenic manipulations in living organisms. Through protein engineering and in vivo functional tests, we report here variants of the Q-system transcriptional activator, including QF2, for driving strong and ubiquitous expression in all Drosophila tissues. Our QF2, Gal4QF and LexAQF chimeric transcriptional activators substantially enrich the toolkit available for transgenic regulation in Drosophila melanogaster.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Brand, A.H. & Perrimon, N. Development 118, 401–415 (1993).
Lai, S.-L. & Lee, T. Nat. Neurosci. 9, 703–709 (2006).
Potter, C.J., Tasic, B., Russler, E.V., Liang, L. & Luo, L. Cell 141, 536–548 (2010).
Silies, M. et al. Neuron 79, 111–127 (2013).
Pérez-Garijo, A., Fuchs, Y. & Steller, H. eLife 2, e01004 (2013).
Mann, K., Gordon, M.D. & Scott, K. Neuron 79, 754–765 (2013).
Wei, X., Potter, C.J., Luo, L. & Shen, K. Nat. Methods 9, 391–395 (2012).
Gohl, D.M. et al. Nat. Methods 8, 231–237 (2011).
Tsuji, G. et al. Mol. Microbiol. 38, 940–954 (2000).
Baum, J.A., Geever, R. & Giles, N.H. Mol. Cell. Biol. 7, 1256–1266 (1987).
Hidalgo, P. et al. Genes Dev. 15, 1007–1020 (2001).
Walters, K.J., Dayie, K.T., Reece, R.J., Ptashne, M. & Wagner, G. Nat. Struct. Mol. Biol. 4, 744–750 (1997).
Kraulis, P.J., Raine, A.R.C., Gadhavi, P.L. & Laue, E.D. Nature 356, 448–450 (1992).
Marmorstein, R., Carey, M., Ptashne, M. & Harrison, S.C. Nature 356, 408–414 (1992).
Ma, J. & Ptashne, M. Cell 48, 847–853 (1987).
Gill, G. & Ptashne, M. Nature 334, 721–724 (1988).
Mondal, K. et al. J. Mol. Biol. 370, 939–950 (2007).
Wilder, E.L. & Perrimon, N. Development 121, 477–488 (1995).
Freeman, M. Cell 87, 651–660 (1996).
Kramer, J.M. & Staveley, B.E. Genet. Mol. Res. 2, 43–47 (2003).
Kelley, L.A. & Sternberg, M.J.E. Nat. Protoc. 4, 363–371 (2009).
Zhang, L., Bermingham-McDonogh, O., Turcotte, B. & Guarente, L. Proc. Natl. Acad. Sci. USA 90, 2851–2855 (1993).
Zdobnov, E.M. & Apweiler, R. Bioinformatics 17, 847–848 (2001).
Pfeiffer, B.D. et al. Genetics 186, 735–755 (2010).
Lee, T. & Luo, L. Neuron 22, 451–461 (1999).
Sharma, Y., Cheung, U., Larsen, E.W. & Eberl, D.F. Genesis 34, 115–118 (2002).
Pfeiffer, B.D. et al. Proc. Natl. Acad. Sci. USA 105, 9715–9720 (2008).
Wu, J.S. & Luo, L. Nat. Protoc. 1, 2110–2115 (2006).
Benzer, S. Proc. Natl. Acad. Sci. USA 58, 1112–1119 (1967).
Gao, X.J. et al. Curr. Biol. 23, 1163–1172 (2013).
Acknowledgements
We thank S. Djuranovic (Johns Hopkins University), A. Radhakrishnan and A. Schuller for S2 cells and training in S2 cell culturing and luciferase assays, B. Smith for help with scanning electron microscopy, T. Shelley for manufacturing the olfactometer setup, J. Simpson (Janelia Farm Research Campus) for an n-Synaptobrevin promoter containing construct, and C.-C. Lin, B. Akitake, R. Reed and Center for Sensory Biology members for discussions and suggestions. This work was supported by grants from the Whitehall Foundation (C.J.P.), US National Institutes of Health (R01DC013070, C.J.P.; R01NS079584, M.N.W., R01 DC005982, L.L.) and Howard Hughes Medical Institute (L.L.).
Author information
Authors and Affiliations
Contributions
C.J.P. and O.R. conceived of the project and designed most of the experiments. S.L. and M.N.W. designed the daily activity and sleep tests. O.R., E.M., S.L. and C.J.P. performed the experiments. D.L. performed embryo injections and generated most of the transgenic animals. L.L. provided reagents and suggestions. O.R. and C.J.P. wrote the manuscript with feedback from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 In vitro and in vivo activities of additional QF variants.
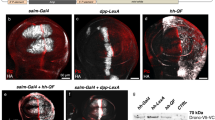
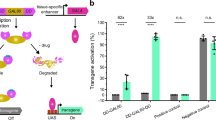
This figure characterises the additional QF variants generated to find the region responsible for QF toxicity. Some variants yielded insufficient transcriptional activity (QFa,c,e,g-k), some were either lethal in vivo (QFd), or exhibited substantial (QFf) or slight (QFb) in vivo toxicity. QFl is an efficient and non-toxic LexAQF chimera, but since a smaller and stronger alternative exists (LexAQF, Fig. 1) that showed similar activity, QFl was not characterized further. (a) Schematics of GAL4, QF and twelve experimental transcriptional activators. BD, DNA binding domain; MD, middle domain; AD, activation domain. Vertical hatching indicates Zn2-Cys6 zinc finger motifs, diagonal hatchings mark dimerization domains. Numbers indicate amino acid position. Constructs are drawn to the same scale. Construct QFb is structurally identical to GAL4QF (Fig. 1), apart from 4 amino acid changes in the C-terminus of the protein (see Online Methods). Construct QFe was made by replacing the C-terminal 22 amino acids of QF with four lysines. This change also likely affected the QS binding site. (b) The transcriptional activity (black bars) of QF variants were investigated by transfecting S2 cells with actin-QFX plasmid, firefly luciferase reporter plasmid (pLexAop-luc2, pQUAS-luc2 or pUAS-luc2), Renilla luciferase plasmid (pAC-hRluc) for normalization, and pBluescript (pBS-KS) plasmid for loading control. For QS repression assays (grey bars), Bluescript plasmid was replaced with pAC-QS plasmid and co-transfected together with actin-QFX, firefly and Renilla luciferase plasmids. QForig is the original Neurospora QF codon sequence from Ref.. QF in this table, and for all variants, has been re-codonized to yield average Drosophila expression levels (see Online methods). Numbers in bars indicate the number of independent repeats. Error bars are SEM. Luciferase activity is shown on a log scale. (c) Pan-neuronal in vivo expression of constructs driven by neuronal synaptobrevin (nsyb) promoter at 25 ˚C. Left column shows mCD8-GFP expression in third-instar larvae (scale bar, 100 μm), middle column shows mCD8-GFP expression in adult brain (nc82, magenta; GFP, green; scale bar, 50 μm), and the right column nuclear lacZ expression in adult brains (elav, red; lacZ, green; nc82, blue). (d) Larval and adult expression of LexAop-mCD8:GFP, driven by the nsyb-QFl construct. Right panel, tub-QS suppression of QFl activity. (e) Quantification of LacZ expression (see Online Methods). Numbers in bars indicate the number of brains for each condition. Error bars are SEM.
Supplementary Figure 2 Quantification of GFP expression driven by nSyb lines at 18 °C, 25 °C and 29 °C.
This experiment is analogous to the one shown in Fig. 1f, but instead of nuclearLacZ reporter, we used a membrane-bound GFP reporter (n=5 for all conditions). Flies were reared at the indicated temperature from embryonic stage until 4 days post-eclosion. This experiment allows us to investigate the possible variability of expression levels due to the nature of the reporter transgene. See Online Methods for details of quantification procedure.
Supplementary Figure 3 Quinic acid–induced disinhibition of expression.
(a) Third instar larvae that express GFP pan-neuronally (left) have GFP expression suppressed by tub-QS (middle) and this suppression is inhibited by raising animals in standard fly medium supplemented by 0.1 g of quinic acid (QA) per 10 ml of food (right). All larvae are imaged at the same settings. Genotypes are indicated below corresponding images. Scale bar, 0.3 mm. (b) Brains of adult flies were immunostained for nc82 (magenta) and GFP (green) and imaged at the same confocal settings. To observe the effect of feeding QA to adult flies, flies were raised to adulthood on standard fly medium and transferred for 3 days into vials that contained 0.6 g of quinic acid (QA, see Online Methods for details). Scale bar, 50 μm.
Supplementary Figure 4 Coexpression of nsyb transactivator driver lines results in viable flies that exhibit robust transgene expression.
Shown are brains of flies that carry two nsyb driver lines, one of which drives pan-neuronal expression of nuclear lacZ (red), and the other drives independent pan-neuronal expression of a membrane-bound GFP (green). Brains are counter-stained with nc82 (blue). nsyb-QF2 and nsyb-QF2w lines both drive expression of QUAS effectors, making it impossible to use these two drivers independently in the same cell. The same applies for nsyb-GAL4 and nsyb-GAL4QF lines which both activate UAS effectors. All tested binary combinations yielded healthy animals, suggesting that at least two independent binary systems can be simultaneously utilized in neuronal tissues. Scale bar, 50 μm.
Supplementary Figure 5 Activity of ubiquitously driven QF2, QF2w, GAL4QF and LexAQF in non-neuronal tissue.
(a) Expression patterns of actin-GAL4QF, tubulin-QF2 and tubulin-QF2w, visualised with a UAS/QUAS-nuclacZ reporter and compared to expression patterns of direct driver tub-nucLacZ (Bloomington stock #29726), actin-GAL4 (Bloomington stock #3954), and tubulin-GAL4 (Bloomington stock #5138). We did not investigate the activity of actin-LexAQF in the same way due to the unavailability of a LexAop-nuclacZ reporter. Tubulin-nucLacZ expression serves as a control for binary system independent expression achievable by the tubulin promoter. Scale bar, 50 μm. (b) Expression patterns represented by ubiquitous GALQF, QF2, QF2w and LexAQF activity in larval body wall muscles. Each driver line was crossed to a corresponding strong hexameric mCherry reporter, as indicated on the figure panel. Third-instar larvae were dissected using the "larval fillet" preparation and imaged directly, without immunostaining, under identical illumination and image acquisition parameters for 150 ms. Wild-type control larvae were imaged for 1000 ms and produced no discernible image. Scale bar, 300 μm. (c) Expression and QS-mediated repression of GALQF, QF2, QF2w and LexAQF activity in larval imaginal discs. Discs from larva that carried a driver and a reporter line (actin-GAL4QF + 5xUAS-mtdt-3HA, actin-LexAQF + 13xLexAop2-6xmcherry-HA, tubulin-QF2 + 5xQUAS-mtdt-3HA or tubulin-QF2w + 5xQUAS-mtdt-3HA) are shown in columns 1, 3, 5 and 7, correspondingly. Columns to the right of these (2, 4, 6 and 8) show imaginal discs from larvae that also carried a tubulin-QS transgene. All discs were immunostained for the HA epitope and counterstained with DAPI using the same protocol (see Online Methods). Weak residual signal in actin-LexAQF + 13xLexAop2-6xmcherry-HA+tub-QS (fourth column) is observed in peripodial membrane cells. Scale bar, 50 μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–5 and Supplementary Note (PDF 4901 kb)
Rights and permissions
About this article
Cite this article
Riabinina, O., Luginbuhl, D., Marr, E. et al. Improved and expanded Q-system reagents for genetic manipulations. Nat Methods 12, 219–222 (2015). https://doi.org/10.1038/nmeth.3250
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3250
This article is cited by
-
A genetically encoded sensor for visualizing leukotriene B4 gradients in vivo
Nature Communications (2023)
-
A non-canonical Raf function is required for dorsal–ventral patterning during Drosophila embryogenesis
Scientific Reports (2022)
-
Transgenic Anopheles mosquitoes expressing human PAI-1 impair malaria transmission
Nature Communications (2022)
-
Enhanced regulation of prokaryotic gene expression by a eukaryotic transcriptional activator
Nature Communications (2021)
-
Intravital imaging strategy FlyVAB reveals the dependence of Drosophila enteroblast differentiation on the local physiology
Communications Biology (2021)