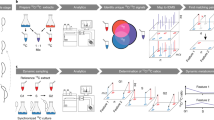
Abstract
Metabolic systems are often the first networks to respond to environmental changes, and the ability to monitor metabolite dynamics is key for understanding these cellular responses. Because monitoring metabolome changes is experimentally tedious and demanding, dynamic data on time scales from seconds to hours are scarce. Here we describe real-time metabolome profiling by direct injection of living bacteria, yeast or mammalian cells into a high-resolution mass spectrometer, which enables automated monitoring of about 300 compounds in 15–30-s cycles over several hours. We observed accumulation of energetically costly biomass metabolites in Escherichia coli in carbon starvation–induced stationary phase, as well as the rapid use of these metabolites upon growth resumption. By combining real-time metabolome profiling with modeling and inhibitor experiments, we obtained evidence for switch-like feedback inhibition in amino acid biosynthesis and for control of substrate availability through the preferential use of the metabolically cheaper one-step salvaging pathway over costly ten-step de novo purine biosynthesis during growth resumption.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Buescher, J.M. et al. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335, 1099–1103 (2012).
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012).
Link, H., Kochanowski, K. & Sauer, U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat. Biotechnol. 31, 357–361 (2013).
Xu, Y.-F., Amador-Noguez, D., Reaves, M.L., Feng, X.-J. & Rabinowitz, J.D. Ultrasensitive regulation of anapleurosis via allosteric activation of PEP carboxylase. Nat. Chem. Biol. 8, 562–568 (2012).
Doucette, C.D., Schwab, D.J., Wingreen, N.S. & Rabinowitz, J.D. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 7, 894–901 (2011).
Chubukov, V., Gerosa, L., Kochanowski, K. & Sauer, U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 12, 327–340 (2014).
Yanes, O. et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 6, 411–417 (2010).
Jain, M. et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044 (2012).
Fuhrer, T., Heer, D., Begemann, B. & Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 83, 7074–7080 (2011).
Potrykus, K. & Cashel, M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51 (2008).
Navarro Llorens, J.M., Tormo, A. & Martínez-García, E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34, 476–495 (2010).
Taymaz-Nikerel, H. et al. Changes in substrate availability in Escherichia coli lead to rapid metabolite, flux and growth rate responses. Metab. Eng. 16, 115–129 (2013).
Brauer, M.J. et al. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA 103, 19302–19307 (2006).
Yoshida, H., Yamamoto, H., Uchiumi, T. & Wada, A. RMF inactivates ribosomes by covering the peptidyl transferase centre and entrance of peptide exit tunnel. Genes Cells 9, 271–278 (2004).
Bujara, M., Schümperli, M., Pellaux, R., Heinemann, M. & Panke, S. Optimization of a blueprint for in vitro glycolysis by metabolic real-time analysis. Nat. Chem. Biol. 7, 271–277 (2011).
Orth, J.D. et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 7, 535 (2011).
Rabinowitz, J.D. & Kimball, E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79, 6167–6173 (2007).
Buescher, J.M., Moco, S., Sauer, U. & Zamboni, N. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal. Chem. 82, 4403–4412 (2010).
Chiang, B.-Y. et al. Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation. J. Biol. Chem. 285, 25345–25353 (2010).
Reeve, C.A., Bockman, A.T. & Matin, A. Role of protein degradation in the survival of carbon-starved Escherichia coli and Salmonella typhimurium. J. Bacteriol. 157, 758–763 (1984).
Yuan, J., Fowler, W.U., Kimball, E., Lu, W. & Rabinowitz, J.D. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat. Chem. Biol. 2, 529–530 (2006).
Akashi, H. & Gojobori, T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 99, 3695–3700 (2002).
Milo, R., Jorgensen, P., Moran, U., Weber, G. & Springer, M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 38, D750–D753 (2010).
Goyal, S., Yuan, J., Chen, T., Rabinowitz, J.D. & Wingreen, N.S. Achieving optimal growth through product feedback inhibition in metabolism. PLoS Comput. Biol. 6, e1000802 (2010).
Koshland, D.E. Switches, thresholds and ultrasensitivity. Trends Biochem. Sci. 12, 225–229 (1987).
Pérez-Arellano, I., Rubio, V. & Cervera, J. Dissection of Escherichia coli glutamate 5-kinase: functional impact of the deletion of the PUA domain. FEBS Lett. 579, 6903–6908 (2005).
Rinas, U., Hellmuth, K., Kang, R., Seeger, A. & Schlieker, H. Entry of Escherichia coli into stationary phase is indicated by endogenous and exogenous accumulation of nucleobases. Appl. Environ. Microbiol. 61, 4147–4151 (1995).
Deo, S.S., Tseng, W.C., Saini, R., Coles, R.S. & Athwal, R.S. Purification and characterization of Escherichia coli xanthine-guanine phosphoribosyltransferase produced by plasmid pSV2gpt. Biochim. Biophys. Acta 839, 233–239 (1985).
Zhou, G., Charbonneau, H., Colman, R.F. & Zalkin, H. Identification of sites for feedback regulation of glutamine 5-phosphoribosylpyrophosphate amidotransferase by nucleotides and relationship to residues important for catalysis. J. Biol. Chem. 268, 10471–10481 (1993).
Fridman, A. et al. Cell cycle regulation of purine synthesis by phosphoribosyl pyrophosphate and inorganic phosphate. Biochem. J. 454, 91–99 (2013).
Skaper, S.D., Willis, R.C. & Seegmiller, J.E. Intracellular 5-phosphoribosyl-1-pyrophosphate: decreased availability during glutamine limitation. Science 193, 587–588 (1976).
Shimaoka, M., Takenaka, Y., Kurahashi, O., Kawasaki, H. & Matsui, H. Effect of amplification of desensitized purF and prs on inosine accumulation in Escherichia coli. J. Biosci. Bioeng. 103, 255–261 (2007).
Madar, D. et al. Promoter activity dynamics in the lag phase of Escherichia coli. BMC Syst. Biol. 7, 136 (2013).
Itzkovitz, S., Blat, I.C., Jacks, T., Clevers, H. & van Oudenaarden, A. Optimality in the development of intestinal crypts. Cell 148, 608–619 (2012).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Christen, S. & Sauer, U. Intracellular characterization of aerobic glucose metabolism in seven yeast species by 13C flux analysis and metabolomics. FEMS Yeast Res. 11, 263–272 (2011).
Canelas, A.B. et al. Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat. Commun. 1, 145 (2010).
Acknowledgements
We thank T. Hwa, M. Basan and M. Zampieri for discussions and D. Karst and M. Morbidelli (Institute of Chemical and Bioengineering, ETH Zurich, Zurich, Switzerland) for providing hybridoma cell cultures.
Author information
Authors and Affiliations
Contributions
H.L., T.F., L.G., N.Z. and U.S. designed the project, analyzed results and wrote the manuscript. H.L. and T.F. performed experiments. H.L. performed simulations.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Real-time metabolome profiling of Saccharomyces cerevisiae.
Shown are fold change of ions annotated to citrate, succinate, AMP, reduced glutathione and lactate. Lines are the mean from 3 biological replicates and colored areas indicate standard deviation. A moving average filter was applied to raw data. Oxygen was removed at t = 10 min by flushing cultures with nitrogen.
Supplementary Figure 2 Growth resumption of E. coli.
Optical density at 600 nm of 2 hour carbon starved E. coli. Glucose was added at t = 0 min to a final concentration of 0.6 g l-1.
Supplementary Figure 3 Global dynamics during starvation and growth resumption.
K-means clustering of z-score normalized time profiles of 302 ions. The data set was clustered into a total of k = 10 clusters. Cluster centers are shown as red dots for the starvation phase (0 - 123 min) and blue dots for the glucose phase (123 - 150 min). Black dots represent individual profiles in the cluster. The number of ions in each cluster is indicated above each plot. Clusters are grouped according to the trends in the starvation and glucose phase as shown above each group. Ionization artifacts are caused by glucose ion suppression (cluster 9) and incomplete clearing of the electrospray chamber when injecting every 15 seconds (cluster 6). Time axis is different for the starvation and glucose phase.
Supplementary Figure 4 Amino acid dynamics during starvation and growth resumption.
Intensity of ions annotated to amino acids. Red: starvation; Blue: glucose phase. The exact mz is given in the upper right corner of each graph. Time axis is different for the starvation and glucose phase.
Supplementary Figure 5 Amino acid–degradation pathways in E. coli.
Pathways are adopted from the EcoCyc Database. Class 1 amino acids have decreased or constant concentrations during starvation. Class 2 amino acids accumulate during starvation (see Supplementary Fig. 4).
Supplementary Figure 6 Absolute concentrations of amino acids obtained by LC-MS.
Cells starved for 123 minutes before glucose was added. Black: samples of the whole cell broth. Magenta: samples of the supernatant. Note that scale of the time axis is different for the starvation and glucose phase.
Supplementary Figure 7 Amino acid dynamics during 23 h of starvation.
(a) Absolute concentrations of amino acids in the whole cell broth during 23 hours starvation measured by LCMS. Lines indicate linear regression for the first data points and numbers the slope in µmol/gDW/h. (b) Rate of accumulation obtained from linear regression is plotted against the according amino acid content in proteins (see Supplementary Table 6). The slope of the linear regression represents the net rate of protein degradation.
Supplementary Figure 8 Response of amino acids to chloramphenicol during exit from starvation.
Magenta: Intensity of ions annotated to the indicated amino acids obtained with the real-time system. 50 µM chloramphenicol was added at t = 135 min, dots are ion intensities and lines a moving average filter. Grey: Response during the glucose phase as shown in Supplementary Figure 4.
Supplementary Figure 9 Flux profiling of chloramphenicol-treated cells.
Fraction of unlabeled amino acids in exponentially growing E. coli washed for 30, 60 and 90 sec with U-13C glucose (black). Fraction of unlabeled amino acids in E. coli that were treated for 5 min with chloramphenicol before washing with U-13C glucose and chloramphenicol (red).
Supplementary Figure 10 Simulation results of the purine nucleotide model.
Simulation results of starvation (-5 min - 123 min) and growth resumption at 123 min obtained with the model of purine nucleotide metabolism (lines). Dots are scaled real-time data of xanthine, AMP and ATP.
Supplementary Figure 11 Growth resumption of E. coli cultures after 22 h of glucose starvation.
After 22 hours cells were centrifuged and resuspended in: old starvation medium (black), fresh M9 medium (blue) or fresh M9 medium supplemented with 20 μM isoleucine, leucine, valine, hypoxanthine and xanthine (red). The indicated carbon source was added at t = 0 min and optical density was monitored in 96-well flat transparent plates in a plate reader. Shown are three replicates (thin lines) and the average (bold lines).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Notes 1 and 2 (PDF 3737 kb)
Supplementary Table 1
Functional categories used for pathway enrichment. Assignment was based on the metabolic model iJO1366 (Ref. 16) (XLSX 19 kb)
Supplementary Table 2
Properties of amino acids and ions annotated to amino acids (XLSX 11 kb)
Supplementary Software 1
Matlab serial port interface control scripts (ZIP 20 kb)
Supplementary Software 2
Matlab kinetic model scripts (ZIP 24 kb)
Rights and permissions
About this article
Cite this article
Link, H., Fuhrer, T., Gerosa, L. et al. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat Methods 12, 1091–1097 (2015). https://doi.org/10.1038/nmeth.3584
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3584
This article is cited by
-
UmetaFlow: an untargeted metabolomics workflow for high-throughput data processing and analysis
Journal of Cheminformatics (2023)
-
An automated workflow for multi-omics screening of microbial model organisms
npj Systems Biology and Applications (2023)
-
Stringent response ensures the timely adaptation of bacterial growth to nutrient downshift
Nature Communications (2023)
-
Time-dependent metabolomics uncover dynamic metabolic adaptions in MCF-7 cells exposed to bisphenol A
Frontiers of Environmental Science & Engineering (2023)
-
Microbial storage and its implications for soil ecology
The ISME Journal (2022)