Abstract
Imaging fast cellular dynamics across large specimens requires high resolution in all dimensions, high imaging speeds, good physical coverage and low photo-damage. To meet these requirements, we developed isotropic multiview (IsoView) light-sheet microscopy, which rapidly images large specimens via simultaneous light-sheet illumination and fluorescence detection along four orthogonal directions. Combining these four views by means of high-throughput multiview deconvolution yields images with high resolution in all three dimensions. We demonstrate whole-animal functional imaging of Drosophila larvae at a spatial resolution of 1.1-2.5 μm and temporal resolution of 2 Hz for several hours. We also present spatially isotropic whole-brain functional imaging in Danio rerio larvae and spatially isotropic multicolor imaging of fast cellular dynamics across gastrulating Drosophila embryos. Compared with conventional light-sheet microscopy, IsoView microscopy improves spatial resolution at least sevenfold and decreases resolution anisotropy at least threefold. Compared with existing high-resolution light-sheet techniques, IsoView microscopy effectively doubles the penetration depth and provides subsecond temporal resolution for specimens 400-fold larger than could previously be imaged.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
05 November 2015
In the version of this article initially published online, Supplementary Video 5 was incorrectly labeled. The error has been corrected as of 5 November 2015.
References
Winter, P.W. & Shroff, H. Faster fluorescence microscopy: advances in high speed biological imaging. Curr. Opin. Chem. Biol. 20, 46–53 (2014).
Keller, P.J. & Ahrens, M.B. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 85, 462–483 (2015).
Chen, B.C. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014).
Wu, Y. et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol. 31, 1032–1038 (2013).
Lemon, W.C. et al. Whole-central nervous system functional imaging in larval Drosophila. Nat. Commun. 6, 7924 (2015).
Tomer, R., Khairy, K., Amat, F. & Keller, P.J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Krzic, U., Gunther, S., Saunders, T.E., Streichan, S.J. & Hufnagel, L. Multiview light-sheet microscope for rapid in toto imaging. Nat. Methods 9, 730–733 (2012).
Schmid, B. et al. High-speed panoramic light-sheet microscopy reveals global endodermal cell dynamics. Nat. Commun. 4, 2207 (2013).
Planchon, T.A. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417–423 (2011).
Vladimirov, N. et al. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods 11, 883–884 (2014).
Mickoleit, M. et al. High-resolution reconstruction of the beating zebrafish heart. Nat. Methods 11, 919–922 (2014).
Bouchard, M.B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat. Photonics 9, 113–119 (2015).
Ahrens, M.B., Orger, M.B., Robson, D.N., Li, J.M. & Keller, P.J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420 (2013).
Panier, T. et al. Fast functional imaging of multiple brain regions in intact zebrafish larvae using selective plane illumination microscopy. Front. Neural Circuits 7, 65 (2013).
Wolf, S. et al. Whole-brain functional imaging with two-photon light-sheet microscopy. Nat. Methods 12, 379–380 (2015).
Swoger, J., Verveer, P., Greger, K., Huisken, J. & Stelzer, E.H. Multi-view image fusion improves resolution in three-dimensional microscopy. Opt. Express 15, 8029–8042 (2007).
Preibisch, S. et al. Efficient Bayesian-based multiview deconvolution. Nat. Methods 11, 645–648 (2014).
Voie, A.H., Burns, D.H. & Spelman, F.A. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc. 170, 229–236 (1993).
Fuchs, E., Jaffe, J., Long, R. & Azam, F. Thin laser light sheet microscope for microbial oceanography. Opt. Express 10, 145–154 (2002).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E.H.K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H.K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008).
Baumgart, E. & Kubitscheck, U. Scanned light sheet microscopy with confocal slit detection. Opt. Express 20, 21805–21814 (2012).
Preibisch, S., Saalfeld, S., Schindelin, J. & Tomancak, P. Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010).
Temerinac-Ott, M., Ronneberger, O., Nitschke, R., Driever, W. & Burkhardt, H. Spatially-variant Lucy-Richardson deconvolution for multiview fusion of microscopic 3D images. In Proc. IEEE International Symposium on Biomedical Imaging 899–904 (IEEE, 2011).
Chen, T.W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Campbell, R.E. et al. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882 (2002).
Morin, X., Daneman, R., Zavortink, M. & Chia, W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15050–15055 (2001).
Fischler, M.A. & Bolles, R.C. Random sample consensus—a paradigm for model-fitting with applications to image-analysis and automated cartography. Commun. ACM 24, 381–395 (1981).
Acknowledgements
We thank B. Coop and the Janelia ID&F Team for mechanical designs and custom mechanical parts; M. Coleman for custom microscope-operating software; V. Stamataki for help with bead injections in live Drosophila embryos and larvae; E. Betzig, K. Branson and G. Rubin for helpful discussions; M. Ahrens and C.-T. Yang (Janelia Research Campus, Ashburn, Virginia, USA) for the GCaMP6s zebrafish line; E. Wieschaus (Howard Hughes Medical Institute, Princeton University, Princeton, New Jersey, USA) for the Spider-GFP flies; the Janelia Fly Facility (Janelia Research Campus, Ashburn, Virginia, USA) for sharing and maintaining transgenic Drosophila stocks; and the Janelia Vivarium Team for zebrafish animal care. This work was supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
P.J.K. and R.K.C. designed the IsoView microscope. R.K.C. built the IsoView microscope and performed all imaging experiments. F.A. developed the IsoView image-processing framework. Y.W. and W.C.L. prepared Drosophila specimens for IsoView imaging. B.H. prepared zebrafish specimens for IsoView imaging. P.J.K. conceived of the research, supervised the project and wrote the paper, with contributions from R.K.C.
Corresponding authors
Ethics declarations
Competing interests
P.J.K. filed a provisional US patent application for IsoView microscopy on October 9, 2013, and P.J.K. and R.K.C. filed a nonprovisional US patent application on October 8, 2014 (application number 14/509,331).
Integrated supplementary information
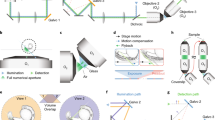
Supplementary Figure 1 Optical design of the IsoView light-sheet microscope.
IsoView illumination arms: Beams from multiple lasers (488 nm, 515 nm, 561 nm, 594 nm), led through single mode fibers (SMF), are collimated by fiber collimators (FC) and directed through illumination filter wheels (IFW) and shutters (S). Lens pairs (L1 and L2) de-magnify the beams to attain the intended Rayleigh length of the weakly focused beams in sample space. De-magnified beams are then scanned by dual-axis galvanometer scanners (XY Galvo) to rapidly generate scanned light sheets as well as to laterally translate light sheets in sample space. The planar scanned fields (dotted lines) generated by the f-theta scan lenses (fθ) are reimaged onto the sample plane by the illumination tube lenses (ITL) and objectives (Obj1, Obj2, Obj3 and Obj4), via reflection from dichroic beam splitters (DBS) en route to sample space.
IsoView detection arms: The emitted fluorescence signal collected from the focal planes of the objectives orthogonal to the light sheets is transmitted by dichroic beam splitters located in infinity space between detection tube lenses (DTL) and objectives. The objectives and detection tube lenses image the fluorescent signal emitted by the specimen onto the sCMOS camera chips, following transmission through the detection filter wheels (DFW). Depth-sectioned images are acquired by linearly translating the objectives mounted atop linear piezos (not shown) and synchronously translating the light sheets laterally to match the focal planes of the objectives collecting the emitted fluorescence light.
We note that, in order to reduce the footprint of the microscope, X and Y scan mirrors in the IsoView illumination arms are not conjugated. Depending on which scan mirror is positioned in the focal plane of the f-theta lens, this arrangement is thus non-telecentric for the respective other scan direction. The implications of this design choice are briefly summarized below.
Case 1: When beam scanning is non-telecentric in the direction that sweeps the Gaussian beam over the camera field, the angle between beam and optical axis is at maximum 1.8 degrees (measured across the full field-of-view, i.e. +/- 400 µm). For a Drosophila-sized specimen, this amounts to a vertical beam offset of 6.3 µm at the two ends of the specimen. Since the size of the confocal slit typically used in IsoView mode 2 is 29.3 µm, the slit is large enough to collect all ballistic photons and there is in principle no impact on image quality.
Case 2: When beam scanning is non-telecentric in the direction that sweeps the light sheet across the sample volume, the angle between light sheet and focal plane is at maximum 0.8 degrees across a 400-µm-deep volume. At the end points of a 200-µm-wide volume this corresponds to a maximum shift of +/- 1.4 µm between light sheet and detection plane. For comparison, the light sheet waist is typically on the order of 6.3 µm.
Supplementary Figure 2 IsoView two-color imaging.
IsoView schematic as shown in Supplementary Fig. 1, but indicating the two different wavelength configurations used for rapid four-view, two-color imaging in IsoView mode 3 (Fig. 1d). The microscope quickly alternates between configurations (a) and (b), in which the specimen is illuminated with two different laser wavelengths and two different spectral bands are imaged along orthogonal axes. Color assignments in the four illumination and detection arms are flipped in orthogonal arms when switching between configurations (a) and (b). Completing a full volumetric imaging cycle with both wavelength configurations produces four-view image data sets for both spectral channels.
Supplementary Figure 3 Technical drawings of IsoView custom optical components.
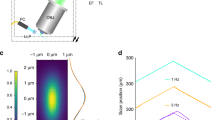
(a) The IsoView microscope uses three types of custom optical components: objectives for both light-sheet illumination and fluorescence detection, f-theta scan lenses and detection tube lenses. These components have been jointly designed and modeled for optimal combined performance in the illumination and detection arms of the IsoView microscope, minimizing spherical and chromatic aberrations and maximizing telecentricity. The most critical custom components are the objectives, which offer (1) diffraction-limited performance over a wide wavelength range (435-950 nm) for multi-color imaging, (2) small telecentric error (< 0.1º) for efficient confocal line scanning, (3) large field-of-view (800 µm) and long working distance (3.09 mm) for imaging large specimens using various specimen preparation techniques, (4) a light-weight design (361 g) for fast volumetric imaging with objective piezo positioners, (5) a cone angle smaller than 90º, which allows simultaneous use of four objectives aimed at a common focal point in a perpendicular arrangement, and (6) a numerical aperture (0.714) close to the theoretical maximum given the geometrical constrains (Supplementary Fig. 5). Higher numerical apertures can be achieved by sacrificing field-of-view or volumetric scan range, but apertures are generally constrained in four-view imaging by the requirement of an objective cone angle below 90º. (b) Technical drawings of the custom optical components shown in a. (c) System PSFs measured with IsoView custom objectives using fluorescent beads located at the surface of an agarose cylinder. Two representative examples are shown. (d) Gaussian fits and respective three-dimensional FWHM measurements of the IsoView system PSF. When imaging eGFP fluorescence using a 525/50-nm band-pass filter, the average emission wavelength is λ = 0.519 µm. Thus, measured lateral resolution is in good agreement with the theoretical Abbe limit of 0.44 µm (0.61 × λ / 0.714).
Supplementary Figure 4 Implementation of the IsoView light-sheet microscope.
(a) Photograph of the core of the IsoView light-sheet microscope. (b) Photograph of the core of the IsoView light-sheet microscope during four-view, two-color imaging (IsoView mode 3, see Fig. 1d) with 488 nm and 561 nm excitation wavelengths. The snapshot shows the microscope during the first phase of the imaging cycle; in the second phase, wavelength assignments in illumination and detection arms are switched in order to acquire the complementary set of colors and views. Together, phases one and two yield four-view data sets for both spectral bands.
Supplementary Figure 5 Geometrical considerations in IsoView objective design.
(a) Schematic top view of the four-objective arrangement in the IsoView microscope. When positioning all objectives symmetrically around the center of the sample chamber, the minimal distance between neighboring objectives is approximately 700 µm. WD = working distance. (b) Schematic side view of an IsoView objective. Black lines indicate the geometrical outline of the objective nose piece. Magenta, green and blue lines mark geometrical distances and angles. Numerical aperture (0.714) is close to the theoretical maximum, considering the geometrical constraints imposed by a field-of-view of 800 µm, a working distance of 3.09 mm (shown in magenta) and a piezo scan range of 800 µm. As illustrated in panels c and d, the minimal distance between neighboring objectives is 440 µm or 150 µm when imaging a volume of 800 × 800 × 800 µm3 in IsoView modes 1 or 2, respectively. (c) Schematic top view of the four-objective arrangement in the IsoView microscope during an imaging experiment performed in IsoView mode 1 (sequential four-view imaging). The snapshot represents the microscope state at the end of a volumetric imaging sequence. When imaging a volume of 800 × 800 × 800 µm3 in this mode, the minimal distance between neighboring objectives is 440 µm. Black arrows next to objective labels indicate objective movement directions during the volume scan. (d) As in c but for IsoView mode 2 (simultaneous four-view imaging). In this scenario, the minimal distance between neighboring objectives is 150 µm when imaging a volume of 800 × 800 × 800 µm3. Black arrows next to objective labels indicate objective movement directions during the volume scan.
Supplementary Figure 6 IsoView point-spread functions and multiview deconvolution.
(a) Lateral and axial dimensions of anisotropic PSFs in views 1 and 2 are flipped with respect to those of views 3 and 4. Improved resolution and isotropy are achieved by combining all views using three-dimensional multi-view image deconvolution. (b) Processing time required to perform four-view image deconvolution with the IsoView image deconvolution software, shown as a function of three-dimensional image size and for four different numbers of iterations of the Lucy-Richardson algorithm (20, 40, 80 and 160). Processing was performed on a single computer workstation equipped with four Nvidia K20x Tesla GPUs. Image size is represented by the voxel count in each single-view image stack. The corresponding computation time measurements also inform about processing time requirements for time-lapse data sets with an arbitrary number of time points. For example, processing a four-view data set with 10243 voxels per view and 16-bit image depth (i.e. 8 GB of image data per time point) takes 128 seconds for 40 Lucy-Richardson iterations, the typical setting used in our study. Thus, processing a typical 10 TB time-lapse data set (1,250 time points, with 8 GB of image data per time point) takes 45.7 hours of GPU time, including I/O operations. (c) Top: Experimental PSFs for views 1-4 (“PSF 1-4”) and final PSF after multi-view deconvolution (“Post-fusion PSF”). Fluorescently labeled beads were imaged at the center of a 1.5-mm-thick agarose cylinder. Bottom: Gaussian fits and FWHM measurements for the PSFs shown above. Post-fusion PSF analysis considers diagonal dimensions of the y-z image plane (see also Swoger et al. 2007, Optics Express). Scale bars, 2 µm (c).
Supplementary Figure 7 IsoView PSFs in a live Drosophila embryo.
(a) Ventral- and lateral-view maximum-intensity projections of a stage 17 Drosophila embryo expressing the calcium indicator GCaMP6s throughout the nervous system. The embryo was injected with fluorescent beads to assess IsoView point spread functions (PSFs) in vivo at different depths in the embryo. Four bead sites are marked by green circles, including two sites in more superficial regions (1 and 2) and two sites at the center of the embryo (3 and 4), i.e. in the deepest and optically most challenging region of the embryo. (b) Left: Experimentally measured PSFs in raw views (labeled “Lateral view PSF” and “Ventral view PSF”, representing the microscope’s orthogonal optical axes) and the final PSF resulting after multi-view deconvolution (“Post-fusion PSF”). The fluorescent bead at site 4 represents a worst-case scenario with respect to the associated imaging challenges: not only is the bead located in the deepest part of the embryo, but it is also found at a location between brain lobes and ventral nerve cord that it difficult to access by the IsoView detection arms aligned with the dorso-ventral axis. Consequently, this view provides the lowest signal-to-noise ratio across all examples. Right: Gaussian fits and respective FWHM measures of PSFs for all views and spatial dimensions. Chi-squared (χ2) residuals for all fits are between 0.0062 and 0.69. Statistics are based on distributions with 16 degrees of freedom (for x-y dimensions in lateral view PSFs, x-z dimensions in ventral view PSFs, all dimensions in post-fusion PSFs) or 42 degrees of freedom (for z dimension in lateral view PSFs, y dimension in ventral view PSFs). Thus, using Pearson's chi-squared test, the probability p of the Gaussian fits to represent poor fits is below 10-10 in all cases. When comparing signal strengths at locations near the surface of the embryo (< 20 µm depth) to those at locations near the center of the embryo (100 µm depth), bead fluorescence intensities decrease on average by a factor of 5 in the raw views and by a factor of 6 in the multi-view deconvolved IsoView data. (c) y-z images and Gaussian fits along diagonal dimensions for the post-fusion PSFs shown in (b). Scale bars, 2 µm (b,c), 50 µm (a).
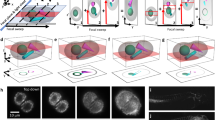
Supplementary Figure 8 Comparison of four-view and orthogonal dual-view imaging (overview).
Side-by-side comparison of multi-view deconvolved image data obtained with orthogonal four-view imaging (IsoView) and orthogonal dual-view imaging (following the diSPIM concept developed by Wu et al. 2013, Nature Biotechnology), for a live stage 17 Drosophila embryo expressing mRFP1 in all cell nuclei. The images show a depth series along the lateral axis of the 200-µm-thick embryo. All images were acquired with the IsoView microscope, using either two (diSPIM) or all four (IsoView) of the microscope’s imaging arms. Objectives were facing the dorsal, ventral, lateral-left or lateral-right sides of the embryo, using dorsal and lateral-left views for diSPIM-like dual-view deconvolution. Imaging settings were otherwise identical, including laser power settings, exposure time and spatial sampling. Image deconvolution was performed with identical point spread functions and number of iterations of the Lucy-Richardson algorithm. Supplementary Figs. 9 and 10 provide enlarged views of x-y and x-z image sections for 24 different regions throughout the embryo (marked by white boxes labeled “ROI1” to “ROI24”). Due to light scattering and light absorption by the embryo, orthogonal dual-view imaging captures approximately one quarter of the sample volume at high spatial resolution, whereas orthogonal four-view imaging provides close to complete coverage of the embryo. We note that IsoView achieves this improvement in high-resolution coverage without decreasing temporal resolution, which is essential for system-level functional imaging with calcium indicators (Supplementary Videos 2,3,4) or developmental imaging of fast cellular dynamics in the Drosophila embryo (Supplementary Videos 6 and 7). Scale bar, 50 µm.
Supplementary Figure 9 Comparison of four-view and orthogonal dual-view imaging (x-y sections).
Enlarged views of x-y image sections for the 24 regions marked by white boxes in Supplementary Fig. 8. The images show a side-by-side comparison of multi-view deconvolved image data obtained with orthogonal four-view imaging (IsoView approach) and orthogonal dual-view imaging (diSPIM approach) for a live stage 17 Drosophila embryo expressing mRFP1 in all cell nuclei. All image data were acquired with the IsoView microscope, using either two (diSPIM) or all four (IsoView) of the microscope’s imaging arms. Imaging and multi-view deconvolution settings were otherwise identical. The line profiles next to the image panels represent normalized intensity profiles along the image y-axis at the respective x-locations indicated by the black arrows. Line profiles are shown both for dual-view imaging (magenta) and four-view imaging (blue). Numbers provided next to the line profiles indicate full-width-at-half-maximum (FWHM) measurements for the left-most intensity peak at the respective x-location. The optical axes of the two objectives employed in dual-view imaging were aligned with the dorsoventral and lateral embryonic axes. The 24 regions analyzed in this comparison cover a wide range of illumination and detection path lengths across the embryo, including regions with diSPIM illumination and detection paths up to 60 µm (ROIs 1, 3, 5 and 7), regions with dorsoventral paths >60 µm and lateral paths <60 µm (ROIs 2, 4, 6 and 8), regions with dorsoventral paths <60 µm and lateral paths >60 µm (ROIs 13-15, 17, 18, 20 and 21-24) and regions with dorsoventral and lateral paths >60 µm (ROIs 9, 10-12, 16 and 19). Dual-view imaging provides excellent spatial resolution up to a maximum depth of approximately 60 µm, yielding high-resolution coverage of approximately one quarter of the specimen. Four-view imaging typically achieved identical spatial resolution in these regions, although a quarter of our data points attest to higher resolution in the IsoView data. These differences are likely rooted in the fact that the respective regions are captured in sufficiently high quality by three out of the four views provided by IsoView, thus adding a third source of high-frequency image content. For illumination and detection path lengths exceeding 60 µm, resolution and signal strength in dual-view imaging degrade substnatially and cellular resolution is typically compromised. In contrast, high-resolution coverage of almost the entire embryo is achieved with four-view imaging, consistent with the intuition that opposing views in IsoView should effectively double optical access along both illumination and detection axes. Notably, four-view imaging still resolves neighboring cell nuclei as distinct objects in some the deepest regions of the embryo with optical path lengths close to 100 µm (ROIs 13 and 16). Scale bars, 5 µm.
Supplementary Figure 10 Comparison of four-view and orthogonal dual-view imaging (x-z sections).
Visualization as in Supplementary Fig. 9, but for enlarged views of x-z image sections for the 24 regions marked by white boxes in Supplementary Fig. 8. We note that all image panels represent individual image slices, rather than volume projections, and thus x-z and x-y sections shown here and in Supplementary Fig. 9 only share a subset of cell identities. Scale bars, 5 µm.
Supplementary Figure 11 Fidelity and noise statistics in GCaMP fluorescence traces.
Side-by-side comparison of single-neuron fluorescence traces and noise statistics for single-view image data (a) and multi-view deconvolved IsoView image data (b). All plots show traces for the same time window and for the same soma located in the posterior ventral nerve cord of a stage 17 Drosophila embryo. Across the four single views (a), signal-to-noise ratio is highest in the ventral-view image data, followed by image data from the two lateral views. We note that the VNC neuron cannot be resolved by imaging from the dorsal side. Extracting a fluorescence trace from the dorsal-view image data for the volume corresponding to the soma volume identified in the ventral-view image data yields a very weak signal (30-fold reduced amplitude) with 15-fold reduced signal-to-noise ratio (second row in (a)). Signal fidelity and noise statistics in the multi-view deconvolved IsoView data are comparable to average performance of the respective two best single views. Increasing the number of iterations of the Lucy-Richardson deconvolution algorithm leads to a slight reduction in signal-to-noise ratio (see left vs. right plots in (b)).
Supplementary Figure 12 Comparison of single-view and IsoView deconvolution (functional imaging).
Side-by-side comparison of raw anisotropic image data, single-view deconvolved image data and multi-view deconvolved IsoView image data for optical sections of the specimen shown in Figs. 2 and 3, using the same false-color look-up-table. Single-view and multi-view deconvolution were performed with identical point spread functions and number of iterations of the Lucy-Richardson algorithm. The comparison includes examples from two different deep regions of the nervous system (dorsal region of the ventral nerve cord and center of a brain lobe). Roman numerals identify locations of somas with high GCaMP6s fluorescence. Single-view deconvolution (rows labeled “Deconvolved views 1+2”, “Deconvolved views 3+4”) improved resolution compared to raw image data (rows labeled “Views 1+2”, “Views 3+4”) but is outperformed by multi-view deconvolution (rows labeled “IsoView”). We further note that single-view deconvolution has a stronger tendency of introducing image artifacts related to noise in the raw image data (for the same number of iterations of the Lucy-Richardson algorithm), in particular for those views with respectively lower signal-to-noise ratio. To take full advantage of IsoView image data, we thus recommend combining the four individual views by multi-view deconvolution. Scale bars, 10 µm.
Supplementary Figure 13 Accurate registration and deconvolution during sample movements.
(a) Columns 1-3: Maximum-intensity projections of raw IsoView image data of an early first instar Drosophila larva expressing the calcium indicator GCaMP6s throughout the nervous system. Projections are shown along view axes 1+2 and 3+4, prior to multi-view deconvolution, for consecutive time points in a fast motor sequence. A wave of muscle contractions is traveling through the larva and reaches peak speed at time point t1. Columns 4-5: RGB color overlays of time points t0, t1 and t2 shown in columns 1-3, visualizing the magnitude of tissue movements during this time interval. The panels in column 5 show enlarged views of the anatomical region most strongly affected by these movements (a section of the ventral nerve cord). (b) Side-by-side comparison of raw, anisotropic image data and multi-view deconvolved IsoView image data for an optical section in the region highlighted in (a). All views were processed at the whole-embryo level with the content-based multi-view registration and deconvolution method described in the Online Methods. (c) Side-by-side comparison of intensity profiles along x-, y- and z-axes extracted from raw, anisotropic image data and multi-view deconvolved IsoView image data, respectively. The locations of the line plots within each optical section are indicated in the panels shown in the top row. Numerical results listed next to the raw intensity profiles represent FWHM size measures. Line profiles and FWHM measures demonstrate that IsoView image registration and multi-view deconvolution operate accurately even in the presence of substantial sample motion; in particular, intensity peaks along the x-axis (i.e. in the direction of maximum sample motion) correspond within one pixel (0.4 µm) in individual views and multi-view deconvolved IsoView image data. Scale bars, 5 µm (b,c), 20 µm (a, enlarged views), 50 µm (a, whole-embryo views).
Supplementary Figure 14 Comparison of single-view and IsoView deconvolution (developmental imaging 1).
Side-by-side comparison of raw anisotropic image data, single-view deconvolved image data and multi-view deconvolved IsoView image data for optical sections of the specimen shown in Fig. 5. Single-view and multi-view deconvolution were performed with identical point spread functions and number of iterations of the Lucy-Richardson algorithm. Single-view deconvolution (rows labeled “Deconvolved views 1+2”, “Deconvolved views 3+4”) improves resolution compared to raw image data (rows labeled “Views 1+2”, “Views 3+4”) but is outperformed by multi-view deconvolution (rows labeled “IsoView”). Importantly, single-view deconvolution also fails to recover morphological features that require multiple views to be properly resolved, such as the cell membranes shown in region 2: views 3+4 contribute membrane features laterally demarcating neighboring cells, whereas views 1+2 contribute membrane features at apical and basal ends of cells. We further note that single-view deconvolution has a stronger tendency of introducing image artifacts related to noise in the raw image data (for the same number of iterations of the Lucy-Richardson algorithm), in particular for those views with respectively lower signal-to-noise ratio. To take full advantage of IsoView image data, we thus recommend combining the four individual views by multi-view deconvolution. Scale bars, 10 µm.
Supplementary Figure 15 Comparison of single-view and IsoView deconvolution (developmental imaging 2).
Comparative visualization as in Supplementary Fig. 14, but for y-z image cross-sections taken from image data of a gastrulating Drosophila embryo (Supplementary Video 5). The lower block of image panels (“Region 5”) exemplifies a worst-case scenario for four-view imaging: membranes and cell nuclei are oriented close to a 45-degree angle relative to the microscope’s two principal imaging axes (y- and z-axes). Thus, for the particular sample geometry encountered here, the four views of the IsoView microscope provide less high-frequency content than available for any other sample geometry (see also Swoger et al. 2007, Optics Express). The corresponding y-z image cross-sections illustrate that IsoView is nevertheless capable of substantial improvements in overall resolution and resolution isotropy relative to conventional imaging, resolving sub-cellular features such as plasma membranes also in this challenging scenario. Scale bar, 10 µm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1 and 2, and Supplementary Note 1 (PDF 4744 kb)
Supplementary Data 1
Technical drawings of IsoView custom mechanical components (ZIP 15297 kb)
Supplementary Data 2
Zemax models of IsoView custom optics (ZIP 699 kb)
Animation of the IsoView microscope
The video shows an animation of the IsoView microscope model. Additional information about IsoView microscope design is provided in the Supplementary Materials, including parts and assembly drawings for all mechanical and optical components of the microscope (Supplementary Data 1), Zemax models of all custom optical components (Supplementary Data 2), a microscope parts list including electronics and computational hardware (Supplementary Table 1, Online Methods), and instructions for setting up and aligning the IsoView microscope (Supplementary Note, Supplementary Figures 1, 2, 3, 4, 5). Note: The video is provided in DivX format. A video player and codecs for the DivX format are freely available at http://www.divx.com/en/software/divx. (AVI 28424 kb)
Comparison of conventional and IsoView functional imaging
Rotating maximum-intensity projections of conventional, non-isotropic image data (left and middle panels) and IsoView image data (right panel) of a stage 17 Drosophila embryo expressing the calcium indicator GCaMP6s throughout the nervous system. The first two panels show pairwise combinations of the four raw, non-isotropic views recorded by IsoView (left: 0° and 180° combined views, middle: 90° and 270° combined views). Views recorded along the same optical axis were fused by adaptive blending, following the standard image processing workflow used for SiMView image data sets (Tomer et al. 2012, Nature Methods), and represent spatial resolution and image quality obtained with conventional light-sheet microscopy. The horizontal dimensions along which spatial resolution is high and low, respectively, are flipped in orthogonal views. The panel to the right shows the final, multi-view deconvolved IsoView image data, which combines information from all four views. Spatial resolution as well as resolution isotropy are greatly improved by orthogonal four-view imaging in conjunction with multi-view image deconvolution. Image data were acquired in IsoView mode 2 (simultaneous four-view imaging with phase-shifted confocal detection, see Fig. 1c) and are displayed using gamma correction and a false-color look-up-table (blue to yellow) to reduce the high dynamic range of the raw data (average signal-to-noise ratio: 211 ± 76, mean ± SD, n = 16) for better visibility. Note: The video is provided in DivX format. A video player and codecs for the DivX format are freely available at http://www.divx.com/en/software/divx. (AVI 14968 kb)
IsoView whole-animal functional imaging in larval Drosophila
Lateral (left), dorsoventral (middle) and rotating (right) maximum-intensity projections of a multi-view deconvolved IsoView time-lapse recording of an early Drosophila first instar larva expressing the calcium indicator GCaMP6s throughout the nervous system. Functional imaging was performed at 2 Hz in IsoView mode 1 (pairwise sequential four-view imaging). The images are gamma-corrected and shown using a false-color look-up-table (blue to yellow) to reduce the high dynamic range of the raw data for better visibility. Note: The video is provided in DivX format. A video player and codecs for the DivX format are freely available at http://www.divx.com/en/software/divx. (AVI 48387 kb)
Long-term IsoView whole-animal functional imaging
Dorsoventral (top) and lateral (bottom) maximum-intensity projections of an IsoView long-term time-lapse recording of a stage 17 Drosophila embryo expressing the calcium indicator GCaMP6s throughout the nervous system. The embryo develops into a first instar larva that crawls out of the imaging volume at the end of the recording. Imaging was performed at 2 Hz in IsoView mode 1 (pairwise sequential four-view imaging) over a period of 9 hours, by alternating between 30-min imaging sessions and 30-min breaks to maximize experiment duration while conserving disk space. Over 120,000 single-view image volumes (4.5 million images, 9 terabytes) were acquired for this specimen, resulting in more than 30,000 multi-view deconvolved high-resolution IsoView image stacks. Note: The video is provided in DivX format. A video player and codecs for the DivX format are freely available at http://www.divx.com/en/software/divx. (AVI 24057 kb)
IsoView whole-brain functional imaging in larval zebrafish.
Dorsoventral (left) and rotating (right) maximum-intensity projections of a multi-view deconvolved IsoView time-lapse recording of the brain of a 3-day old larval zebrafish expressing the calcium indicator GCaMP6s throughout its nervous system. Functional imaging was performed at 1 Hz in IsoView mode 1 (pairwise sequential four-view imaging). The video captures several instances of motor-related, large-scale brain activity in the awake animal. For better visibility of the large dynamic range of the image data, images are presented using a false-color look-up-table (blue to yellow). The video is provided in DivX format. A video player and codecs for the DivX format are freely available at http://www.divx.com/en/software/divx. (AVI 35413 kb)
Comparison of conventional and IsoView developmental imaging
Rotating maximum-intensity projections of conventional, non-isotropic image data (left and middle panels) and IsoView image data (right panel) of a stage 5 Drosophila embryo ubiquitously expressing GFP targeted to membranes and RFP targeted to cell nuclei (colors are inverted for better visibility). The first two panels show pairwise combinations of the four raw, non-isotropic views recorded by IsoView (left: 0° and 180° combined views, middle: 90° and 270° combined views). Views recorded along the same optical axis were fused by adaptive blending, following the standard image processing workflow used for SiMView image data sets (Tomer et al. 2012, Nature Methods), and represent spatial resolution and image quality obtained with conventional light-sheet microscopy. The horizontal dimensions along which spatial resolution is high and low, respectively, are flipped in orthogonal views. The panel to the right shows the final, multi-view deconvolved IsoView image data, which combines information from all four views. Spatial resolution as well as resolution isotropy are greatly improved by orthogonal four-view imaging in conjunction with multi-view image deconvolution. Image data were acquired in IsoView modes 2 and 3 (four-view two-color imaging with phase-shifted confocal detection, see Fig. 1c,d) and are displayed using gamma correction to reduce the high dynamic range of the raw data (average signal-to-noise ratio: 132 ± 43, mean ± SD, n = 8) for better visibility. We note that the vertical transition regions visible in the pairwise combined views (at 0° and 90° rotation angles for the middle and left panels, respectively) are a result of linear blending of the two image volumes and imperfect physical coverage of these regions by two-view imaging. Owing to the two-fold better physical coverage of IsoView four-view imaging, such imaging artifacts are avoided in the IsoView reconstruction (right panel). Note: The video is provided in DivX format. A video player and codecs for the DivX format are freely available at http://www.divx.com/en/software/divx. (AVI 14454 kb)
IsoView two-color imaging of Drosophila gastrulation
From left to right: dorsal, ventral, lateral-left, lateral-right and rotating maximum-intensity projections of a multi-view deconvolved IsoView time-lapse recording of a Drosophila embryo ubiquitously expressing GFP targeted to membranes and RFP targeted to cell nuclei (colors are inverted for better visibility). The video shows embryonic development during stages 6-8. Image data were acquired at 4-second intervals in IsoView modes 2 and 3 (four-view two-color imaging with phase-shifted confocal detection, see Fig. 1c,d) and are displayed using gamma correction to reduce the high dynamic range of the raw data for better visibility. Note: The video is provided in DivX format. A video player and codecs for the DivX format are freely available at http://www.divx.com/en/software/divx. (AVI 26387 kb)
Rights and permissions
About this article
Cite this article
Chhetri, R., Amat, F., Wan, Y. et al. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Methods 12, 1171–1178 (2015). https://doi.org/10.1038/nmeth.3632
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3632
This article is cited by
-
A single-cell transcriptomic atlas of complete insect nervous systems across multiple life stages
Neural Development (2022)
-
Standard metadata for 3D microscopy
Scientific Data (2022)
-
Practical considerations for quantitative light sheet fluorescence microscopy
Nature Methods (2022)
-
In toto light sheet fluorescence microscopy live imaging datasets of Ceratitis capitata embryonic development
Scientific Data (2022)
-
Lightsheet optical tweezer (LOT) for optical manipulation of microscopic particles and live cells
Scientific Reports (2022)