Abstract
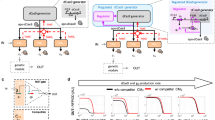
The ability to dynamically manipulate the transcriptome is important for studying how gene networks direct cellular functions and how network perturbations cause disease. Nuclease-dead CRISPR–dCas9 transcriptional regulators, while offering an approach for controlling individual gene expression, remain incapable of dynamically coordinating complex transcriptional events. Here, we describe a flexible dCas9-based platform for chemical-inducible complex gene regulation. From a screen of chemical- and light-inducible dimerization systems, we identified two potent chemical inducers that mediate efficient gene activation and repression in mammalian cells. We combined these inducers with orthogonal dCas9 regulators to independently control expression of different genes within the same cell. Using this platform, we further devised AND, OR, NAND, and NOR dCas9 logic operators and a diametric regulator that activates gene expression with one inducer and represses with another. This work provides a robust CRISPR–dCas9-based platform for enacting complex transcription programs that is suitable for large-scale transcriptome engineering.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Maurano, M.T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Weinhold, N., Jacobsen, A., Schultz, N., Sander, C. & Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165 (2014).
DeRisi, J.L., Iyer, V.R. & Brown, P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686 (1997).
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008).
Loew, R., Heinz, N., Hampf, M., Bujard, H. & Gossen, M. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol 10, 81 (2010).
No, D., Yao, T.P. & Evans, R.M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 93, 3346–3351 (1996).
Doudna, J.A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Qi, L.S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Gilbert, L.A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Maeder, M.L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Cheng, A.W. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171 (2013).
Polstein, L.R. & Gersbach, C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 11, 198–200 (2015).
Nihongaki, Y., Yamamoto, S., Kawano, F., Suzuki, H. & Sato, M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 22, 169–174 (2015).
Zetsche, B., Volz, S.E. & Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142 (2015).
Jusiak, B., Cleto, S., Perez-Piñera, P. & Lu, T.K. Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol. 34, 535–547 (2016).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Liang, F.-S., Ho, W.Q. & Crabtree, G.R. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal. 4, rs2 (2011).
Miyamoto, T. et al. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat. Chem. Biol. 8, 465–470 (2012).
Inoue, T., Heo, W.D., Grimley, J.S., Wandless, T.J. & Meyer, T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods 2, 415–418 (2005).
Levskaya, A., Weiner, O.D., Lim, W.A. & Voigt, C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Kennedy, M.J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010).
Yazawa, M., Sadaghiani, A.M., Hsueh, B. & Dolmetsch, R.E. Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 27, 941–945 (2009).
Liu, H. et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 (2008).
Toettcher, J.E., Gong, D., Lim, W.A. & Weiner, O.D. Light control of plasma membrane recruitment using the Phy-PIF system. Methods Enzymol. 497, 409–423 (2011).
Margolin, J.F. et al. Krüppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 91, 4509–4513 (1994).
Ran, F.A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Nishimasu, H. et al. Crystal structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015).
Kiani, S. et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods 12, 1051–1054 (2015).
Chen, B. et al. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 44, e75 (2016).
Tanenbaum, M.E., Gilbert, L.A., Qi, L.S., Weissman, J.S. & Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Chavez, A. et al. Comparison of Cas9 activators in multiple species. Nat. Methods 13, 563–567 (2016).
Ueguchi-Tanaka, M. et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 (2005).
Miyazono, K. et al. Structural basis of abscisic acid signalling. Nature 462, 609–614 (2009).
Thakore, P.I. et al. Highly specific epigenome editing by CRISPR–Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015).
Nihongaki, Y., Kawano, F., Nakajima, T. & Sato, M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760 (2015).
Hemphill, J., Borchardt, E.K., Brown, K., Asokan, A. & Deiters, A. Optical Control of CRISPR/Cas9 Gene Editing. J. Am. Chem. Soc. 137, 5642–5645 (2015).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Roybal, K.T. et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 164, 770–779 (2016).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell 164, 780–791 (2016).
Kearns, N.A. et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods 12, 401–403 (2015).
Hilton, I.B. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015).
Kiani, S. et al. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–726 (2014).
Nissim, L., Perli, S.D., Fridkin, A., Perez-Pinera, P. & Lu, T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 54, 698–710 (2014).
Acknowledgements
The authors thank the members of the Qi and Lim labs for advice and helpful discussions and the Stanford Shared FACS Facility for technical support. The authors also thank X. Shu (UCSF), F. Zhang (MIT), G. Crabtree (Stanford), T. Inoue (Johns Hopkins), T. Meyer (Stanford), C. Tucker (U of Colorado), and R. Dolmetsch (Stanford) for constructs used in this study. L.S.Q. acknowledges support from the NIH Office of the Director (OD) and the National Institute of Dental & Craniofacial Research (NIDCR). Y.G. acknowledges support from the Stanford Cancer Biology Graduate Program and the NSF GRFP fellowship. X.X. acknowledges the support from the Helen Hay Whitney Foundation postdoctoral fellowship. This work was supported by DP5 OD017887 (L.S.Q.), NIH R01 DA036858 (L.S.Q. and W.A.L.), NIH P50 GM081879 (W.A.L.), and the Howard Hughes Medical Institute (W.A.L.).
Author information
Authors and Affiliations
Contributions
L.S.Q. and W.A.L. conceived of the research; Y.G. and X.X. designed the study; Y.G., X.X., S.W., and E.J.C. performed the experiments; Y.G., X.X., S.W., and L.S.Q. analyzed the data; Y.G., X.X., and L.S.Q. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
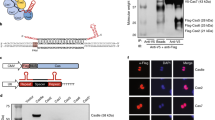
Supplementary Figure 1 Screen of chemical- and light-mediated heterodimerization systems for inducible dCas9 gene activation.
(a) PiggyBac single vector design for expression of dCas9 and effector cassettes. (b) Binding sites of Sp sgTRE3G on the pTRE3G promoter. (c-f) Fluorescence quantification after 48 h induction for HEK293T pTRE3G-EGFP cells transfected with Sp TRE3G and (c) rapamycin-inducible, (d) PHYB/PIF red light-inducible, (e) CRY2PHR/CIBN blue light inducible, or (f) FKF1/GI blue light-inducible VPR-Sp dCas9. The data are displayed as mean ± SD for two (d-f) or four (c) independent transfections performed in technical replicates (n = 4 or 8). (* P < 0.05, ** P < 0.01, *** P < 0.001, n.s. = not significant, Games-Howell post-hoc test following Welch’s ANOVA).
Supplementary Figure 2 Gene activation and repression by direct fusion dCas9s.
(a) Fluorescence quantification for HEK293T pTRE3G-EGFP cells 3 days following transfection with Sp sgTRE3G and VPR-Sp dCas9. (b) Fluorescence quantification for HEK293T pSV40-EGFP cells 6 days following transfection with Sp sgSV40 and KRAB-Sp dCas9. (c) Fluorescence quantification for HEK293T pTRE3G-EGFP cells 3 days following transfection with Sa sgTRE3G and VPR-Sa dCas9. Fold-change activation is indicated in black, while inverse fold-change repression is indicated in magenta. The data are displayed as mean ± SD for four independent transfections performed in technical replicates (n = 8). (* P < 0.05, ** P < 0.01, *** P < 0.001, n.s. = not significant, Welch’s two-sided t test).
Supplementary Figure 3 Gene activation by different configurations of inducible dCas9s.
(a-c) Fluorescence quantification after 48 h induction for HEK293T pTRE3G-EGFP cells stably expressing Sp sgTRE3G and one of two configurations of (a) ABA-inducible, (b) PHYB/PIF red light-inducible, or (c) rapamycin-inducible VP64-Sp dCas9. The data are displayed as mean ± SD for two independent platings of a stable cell line performed in technical replicates (n = 4). †Data collected from a separate experiment consisting of one plating. (d-f) Fluorescence quantification after 48 h induction for HEK293T pTRE3G-EGFP cells transfected with Sp sgTRE3G and one of two configurations of (d) GA-inducible, (e) CRY2PHR/CIBN blue light-inducible, or (f) FKF1/GI blue light-inducible VP64-Sp dCas9. The data are displayed as mean ± SD for two independent transfections performed in technical replicates (n = 4).
Supplementary Figure 4 Gene repression characterization for ABA-inducible KRAB-Sp dCas9.
(a) Binding sites of Sp sgSV40 on the pSV40 promoter. (b) 7 d timecourse for clonal HEK293T pSV40-EGFP cells stably expressing Sp sgSV40 and ABA-inducible KRAB-Sp dCas9. Cells were induced with ABA for 7 d (ON7) or for 4 and 5 d followed by ABA removal (ON4 OFF3 and ON5 OFF2). (c) ABA dose response after 5 d induction for clonal HEK293T pSV40-EGFP cells stably expressing Sp sgSV40 and ABA-inducible KRAB-Sp dCas9. The data are displayed as mean ± SD for two (b) or four (c) independent platings of a stable cell line (n = 2 or 4).
Supplementary Figure 5 Orthogonality of inducible dCas9 components.
(a) Fully orthogonal inducible dCas9 systems require each component of one system to avoid crosstalk to its counterpart in the paired system. (b) Fluorescence quantification after 48 h induction of HEK293T pTRE3G-EGFP cells transfected with Sp sgTRE3G and ABA-inducible or GA-inducible VP64-Sp dCas9. (c) Fluorescence quantification for HEK293T pTRE3G-EGFP cells 72 h following transfection with Sp/Sa sgTRE3G and direct fusion VPR-Sp/Sa dCas9. (d) Fluorescence quantification after 48 h induction of HEK293T pTRE3G-EGFP cells transfected with Sp/Sa sgTRE3G and ABA-inducible VP64-Sp/Sa dCas9. The data are displayed as mean ± SD for two independent transfections performed in technical replicates (n = 4). (* P < 0.05, ** P < 0.01, *** P < 0.001, n.s. = not significant, Games-Howell post-hoc test following Welch’s ANOVA).
Supplementary Figure 6 Independent gene regulation by orthogonal dCas9s.
(a) 2-D flow cytometry contour plots of EGFP and mCherry fluorescence quantification after 5 d induction for experiment shown in Fig. 3b. The plots display data from one of four independent transfections for each condition. (b) 2-D flow cytometry contour plots of CD95 and CXCR4 immunofluorescence quantification after 48 h induction for experiment shown in Fig. 3d. The plots display data from one of four independent transfections for each condition.
Supplementary Figure 7 Two-input logic gates for inducible gene activation and repression.
(a) Fluorescence quantification after 48 h induction for HEK293T pTRE3G-EGFP cells transfected with Sp sgTRE3G and one of two configurations of an AND gate VP64-Sp dCas9. (b-c) Fluorescence quantifications after 5 d induction for HEK293T pSV40-EGFP cells transiently transfected with Sp sgSV40 and (a) the NOR gate or (b) the NAND gate KRAB-Sp dCas9 construct. Fold-change activation is indicated in black, while inverse fold-change repression is indicated in magenta. The data are displayed as mean ± SD for two (a, c) or four (b) independent transfections performed in technical replicates (n = 4 or 8). (* P < 0.05, ** P < 0.01, *** P < 0.001, n.s. = not significant, Games-Howell post-hoc test following Welch’s ANOVA).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1 and 2, Supplementary Notes 1 and 2, and Supplementary Protocol. (PDF 1644 kb)
Rights and permissions
About this article
Cite this article
Gao, Y., Xiong, X., Wong, S. et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods 13, 1043–1049 (2016). https://doi.org/10.1038/nmeth.4042
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4042
This article is cited by
-
Orthogonal inducible control of Cas13 circuits enables programmable RNA regulation in mammalian cells
Nature Communications (2024)
-
A universal deep-learning model for zinc finger design enables transcription factor reprogramming
Nature Biotechnology (2023)
-
Engineering cell-derived extracellular vesicles for gene therapy
Nature Biomedical Engineering (2023)
-
CasTuner is a degron and CRISPR/Cas-based toolkit for analog tuning of endogenous gene expression
Nature Communications (2023)
-
Combinatorial protein dimerization enables precise multi-input synthetic computations
Nature Chemical Biology (2023)