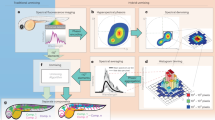
Abstract
Time-lapse imaging of multiple labels is challenging for biological imaging as noise, photobleaching and phototoxicity compromise signal quality, while throughput can be limited by processing time. Here, we report software called Hyper-Spectral Phasors (HySP) for denoising and unmixing multiple spectrally overlapping fluorophores in a low signal-to-noise regime with fast analysis. We show that HySP enables unmixing of seven signals in time-lapse imaging of living zebrafish embryos.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Garini, Y., Young, I.T. & McNamara, G. Cytometry A 69, 735–747 (2006).
Dickinson, M.E., Simbuerger, E., Zimmermann, B., Waters, C.W. & Fraser, S.E. J. Biomed. Opt. 8, 329–338 (2003).
Dickinson, M.E., Bearman, G., Tille, S., Lansford, R. & Fraser, S.E. Biotechniques 31, 1272, 1274–1276, 1278 (2001).
Levenson, R.M. & Mansfield, J.R. Cytometry A 69, 748–758 (2006).
Jahr, W., Schmid, B., Schmied, C., Fahrbach, F.O. & Huisken, J. Nat. Commun. 6, 7990 (2015).
Lansford, R., Bearman, G. & Fraser, S.E. J. Biomed. Opt. 6, 311–318 (2001).
Zimmermann, T. Adv. Biochem. Engin. Biotechnol. 95, 245–265 (2005).
Jolliffe, I. Principal Component Analysis (Wiley, 2002).
Gong, P. & Zhang, A. Geogr. Info. Sci. 5, 52–57 (1999).
Mukamel, E.A., Nimmerjahn, A. & Schnitzer, M.J. Neuron 63, 747–760 (2009).
Fereidouni, F., Bader, A.N. & Gerritsen, H.C. Opt. Express 20, 12729–12741 (2012).
Andrews, L.M., Jones, M.R., Digman, M.A. & Gratton, E. Biomed. Opt. Express 4, 171–177 (2013).
Cutrale, F., Salih, A. & Gratton, E. Methods Appl. Fluoresc. 1, 035001 (2013).
Cranfill, P.J. et al. Nat. Methods 13, 557–562 (2016).
Vermot, J., Fraser, S.E. & Liebling, M. HFSP J. 2, 143–155 (2008).
Trinh, A. et al. Genes Dev. 25, 2306–2320 (2011).
Jin, S.W., Beis, D., Mitchell, T., Chen, J.N. & Stainier, D.Y. Development 132, 5199–5209 (2005).
Livet, J. et al. Nature 450, 56–62 (2007).
Lichtman, J.W., Livet, J. & Sanes, J.R. Nat. Rev. Neurosci. 9, 417–422 (2008).
Pan, Y.A. et al. Development 140, 2835–2846 (2013).
Westerfield, M. The Zebrafish Book (University Oregon Press, 1994).
Megason, S.G. Methods Mol. Biol. 546, 317–332 (2009).
Clayton, A.H., Hanley, Q.S. & Verveer, P.J. J. Microsc. 213, 1–5 (2004).
Redford, G.I. & Clegg, R.M. J. Fluoresc. 15, 805–815 (2005).
Digman, M.A., Caiolfa, V.R., Zamai, M. & Gratton, E. Biophys. J. 94, L14–L16 (2008).
Dalal, R.B., Digman, M.A., Horwitz, A.F., Vetri, V. & Gratton, E. Microsc. Res. Tech. 71, 69–81 (2008).
Fereidouni, F., Reitsma, K. & Gerritsen, H.C. Opt. Express 21, 11769–11782 (2013).
Chen, H., Gratton, E. & Digman, M.A. Microsc. Res. Tech. 78, 283–293 (2015).
Hamamatsu Photonics, K.K. Photomultiplier Technical Handbook (Hamamatsu Photonics K.K., 1994).
Acknowledgements
The authors thank T.V. Truong, C. Arnesano, M. Kitano, S. Restrepo (Translational Imaging Center, University of Southern California) and G.H. Bearman for helpful discussions. This work was supported by grants from the Moore Foundation, the Coulter Foundation and the NIH (R01 HD075605, R01 OD019037). We thank C. Paquette for fish care.
Author information
Authors and Affiliations
Contributions
F.C. and V.T. performed experiments and analyzed the results. J.M.C., L.A.T. and S.E.F. helped in the experimental design and data analysis. V.T. derived equations. F.C. wrote the software. M.S.A. prepared the samples. V.T., F.C., L.A.T., C.-L.C. and S.E.F. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The University of Southern California has filed a provisional patent application (62419075) covering this method.
Integrated supplementary information
Supplementary Figure 1 Comparison of HySP and linear unmixing under different signal-to-noise ratios (SNRs).
(a) TrueColor images of 32 channel datasets of zebrafish labeled with H2B-Cerulean, kdrl:eGFP, Desm-Citrine, Xanthophores, membrane-mCherry as well as Autofluorescence at 458nm and 561nm. The original dataset (SNR 20) was digitally degraded by adding noise and decreasing signal down to SNR 10 and to SNR 5. (b) Normalized spectra used for non-weighted linear unmixing. Spectra were identified on each sample from anatomical regions known to contain only the specific label. For example Xanthophore’s spectrum was collected in dorsal area, H2B-Cerulean’s from fin, kdrl:eGFP’s intramuscularly. Linear unmixing results were optimized over multiple iteration of unmixing regions combinations. The same regions were then used for all three datasets. The same legend and color coding is used through the entire figure. (c) Processed zoomed-in region (box in (a)) for linear unmixing and HySP. The comparison shows three nuclei belonging to muscle fiber. At higher SNR (20 and above) both linear unmixing and HySP results are accurate. Lowering SNR, however, affects the linear unmixing more than the phasor. This can improve unmixing of labels in volumetric imaging of biological samples, where generally SNR decreases with depth and explains the differences in Figure 2e,f, Supp. Figure 6, Supp. Figure 7e,f and Supp. Figure 9. The key advantage of HySP, in this SNR comparison, is the spectral denoising in Fourier space (Supplementary Discussion 3). (e) Intensity profile (dashed arrow in (c)) comparison shows the improvement of HySP at low SNR. Under decreased SNR H2B-Cerulean (cyan) and Desm-Citrine (yellow) (solid arrows in (c)) are consistently identified in HySP while they are partially mislabeled in linear unmixing. For example, some noisy are identified as kdrl:eGFP (green) while, anatomically no vasculature is present in this region of interest.
Supplementary Figure 2 Errors on hyper-spectral phasor plot.
(a) scatter error scales inversely as the square root of the total digital counts. Scatter error also depends on the Poissonian noise in the recording. R-squared statistical method is used to confirm linearity with the reciprocal of square root of counts. The slope is a function of the detector gain used in acquisition showing the counts-to-scatter error dynamic range is inversely proportional to the gain. Lower gains produce smaller scatter error at lower intensity values. The legend is applicable to all parts of the figure. (b) Denoising in the phasor space reduces the scatter error without affecting the location of expected values (ze(n)) on the phasor plot. (c) Denoised scatter error linearly depends on the scatter error without filtering, irrespective of the acquisition parameters. The slope is determined by the filter size (3x3 here). (d) Denoising does not affect normalized shifted-mean errors since the locations of ze(n)’s on the phasor plot remain unaltered due to filtering (d). In this case one filtering was applied.
Supplementary Figure 3 Sensitivity of phasor point.
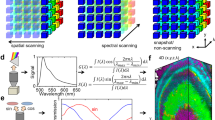
(a,b,c) |Z(n)| remains nearly constant for different imaging parameters. Legend applies to (a,b,c,d,e). (d) Total digital counts as a function of laser power. (e) Proportionality constant in eq. 2 depends on the gain. (f) Relative magnitudes of residuals (R(n)) on phasor plots at harmonics 1 to 16, shows that harmonics n =1 and 2 are sufficient for unique representation of spectral signals.
Supplementary Figure 4 Effect of phasor space denoising on scatter error and shifted-mean error.
(a) Scatter Error as a function of digital counts for different numbers of denoising filters with 3 by 3 mask. Data origin is fluorescein dataset acquired at gain 800 (Supp. Table 2). (b) Scatter Error as a function of number of denoising filters with 3 by 3 mask for different laser powers. (c) Shifted-Mean Error as a function of digital counts for different number of denoising filters with 3 by 3 mask. Data origin is fluorescein dataset acquired at gain 800. (d) Shifted-Mean Error as a function of number of filters with 3 by 3 mask for different laser powers. (e) Relative change of Scatter Error as a function of number of denoising filters applied for different mask sizes. (f) Relative change of Shifted-Mean Error as a function of number of filters applied for different mask sizes.
Supplementary Figure 5 Effect of phasor space denoising on image intensity.
(a,b) HySP processed Citrine channel of a dual labeled eGFP-Citrine sample (132.71um x 132.71um) before and after filtering in phasor space. (c,d) HySP processed eGFP channel of the sample in (a,b) before and after filtering in phasor space. (e) Total intensity profile of the green line highlighted in (a,b,c,d) for different number of denoising filters. Intensity values are not changing. (f) eGFP channel intensity profile of green line highlighted in (a,b,c,d) for different number of denoising filters. (g) Citrine channel intensity profile of green line highlighted in (a,b,c,d) for different number of denoising filters.
Supplementary Figure 6 Phasor analysis for unmixing hyper-spectral fluorescent signals in vivo.
(a) Schematic of the expression patterns of Citrine (skeletal muscles) and eGFP (endothelial tissue) in transgenic zebrafish lines Gt(desm-citrine)ct122a/+ and Tg(kdrl:eGFP) respectively. (b) Conventional optical filter separation for Gt(desm-citrine)ct122a/+ Tg(kdrl:eGFP). Using emission bands on detector of spectrally overlapping fluorophores (eGFP and citrine) cannot overcome the problem of bleed-through of signal in respective channels. Arrows indicate erroneous detection of eGFP or Citrine expressions in the other channel. Scale bar, 200μm. (c) Phasor plots showing spectral fingerprints (scatter densities) for Citrine and eGFP in individually expressed embryo and double transgenic. The individual Citrine and eGFP spectral fingerprints remain preserved in the double transgenic line. (d) Maximum intensity projection images reconstructed by mapping the scatter densities from phasor plot to the original volume. eGFP and Citrine fingerprints can cleanly distinguish the skeletal muscles from interspersed blood vessels (endothelial tissue), within the same anatomical region of the embryo, in both single and double transgenic lines. Scale bar 300μm. Embryos imaged around 72 hours post fertilization. (e,f) HySP analysis outperforms optical separation and linear unmixing in distinguishing spectrally overlapping fluorophores in vivo. (e) Maximum intensity projection images of the region in Tg(kdrl:eGFP);Gt(desm-citrine)ct122a/+ shown in (d) compares the signal for eGFP and Citrine detected by optical separation, linear unmixing and phasor analysis. (f) Corresponding normalized intensity profiles along the width (600 pixels, 553.8μm) of the image integrated over a height of 60 pixels. Correlation values (R) reported for the three cases show the lowest value for HySP analysis, as expected by the mutually exclusive expressions of the two proteins.
Supplementary Figure 7 Autofluorescence identification and removal in phasor space.
(a) Phasor plots showing spectral fingerprints (scatter densities) for citrine, eGFP and autofluorescence allow simple identification of intrinsic signal. (b) Maximum intensity projection images reconstructed by mapping the scatter densities from phasor plot to the original volume. Autofluorescence has a broad fingerprint that can effectively be treated as a fluorescent signature and saved as a channel. Embryos imaged around 72 hours post fertilization.
Supplementary Figure 8 Optical separation of eGFP and citrine.
(a) Spectra of citrine (peak emission 529nm, skeletal muscles) and eGFP (peak emission 509nm, endothelial tissue) measured using confocal multispectral lambda mode in transgenic zebrafish lines Gt(desm-citrine)ct122a/+ and Tg(kdrl:eGFP) respectively. (b) Conventional optical separation (using emission bands on detector) of spectrally close fluorophores (eGFP and citrine) cannot overcome the problem of bleed-through of signal in respective channels. Arrows indicate erroneous detection of eGFP or citrine expressions in the other channel. Scale bar 300μm. (c) Normalized intensity profiles along the line (600 pixels, 553.8μm) in panel (a)
Supplementary Figure 9 Comparison of HySP and linear unmixing in resolving 7 fluorescent signals.
(a) Gray scale images from different optical sections, same as the ones used in Fig 2 (Regions 1-3), comparing the performance of HySP analysis and linear unmixing. (b) Normalized intensity plots for comparison of HySP analysis and linear unmixing. Similar to the corresponding panels in Fig. 2f, the x-axes denote the normalized distance and y-axes in all graphs were normalized to the value of maximum signal intensity among the 7 channels to allow relative comparison. The panels show all intensity profiles for 7 channels in the respective images.
Supplementary Figure 10 Effect of binning on HySP analysis of 7 in vivo fluorescent signals.
(a) The original dataset acquired with 32 channels is computationally binned sequentially to 16, 8 and 4 channels to understand the limits of HySP in unmixing the selected fluorescence spectral signatures. The binning does not produce visible deterioration of the unmixing. White square area is used for zoomed comparison of different bins. (b) Hyper-spectral phasor plots at 458nm and 561nm excitation. Binning of data results in shorter phasor distances between different fluorescent spectral fingerprints. Clusters, even if closer, are still recognizable. (c) Zoomed-in comparison of embryo trunk (box in (a)). Differences for HySP analysis for the same dataset at different binning values are still subtle to the eye. One volume is chosen for investigating intensity profiles (white dashed arrow). (d) Intensity profiles for kdrl:eGFP, H2B-Cerulean, Desm-Citrine and Xanthophores at different binning for summed intensities of a volume of 26.60x0.27x20.00 μm (white dashed arrow (c)). The effects of binning are now visible. For kdrl:eGFP the unmixing is not excessively deteriorated by the binning. Same result for H2B-Cerulean. Desm-Citrine and Xanthophores seems to be more affected by binning. This result suggests that, in our case of zebrafish embryo with 7 separate spectral fingerprints acquired sequentially using two different lasers, it is possible to use 4 bins at the expense of a slight deterioration of the unmixing.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 1–3 and Supplementary Notes 1–6. (PDF 2546 kb)
Supplementary Software 1
HyperSpectral Phasor HySP. MacOSX version 10.11+ (ZIP 58683 kb)
Supplementary Software 2
HyperSpectral Phasor HySP. Windows x64 version (ZIP 179765 kb)
Truecolor rendering of raw data zebrafish with 2 fluorescent and 1 auto-fluorescent labels
Truecolor 32 channel z-stack of Gt(desm-Citrine)ct122aR/+;Tg(kdrl:eGFP) (MP4 6523 kb)
HySP resolved 2-color zebrafish with autofluorescence removal
HySP resolved z-stack of Gt(desm-Citrine)ct122aR/+;Tg(kdrl:eGFP) with auto-fluorescence as a separate channel (MP4 10537 kb)
Truecolor rendering of raw data zebrafish head with 4 fluorescent and 3 auto-fluorescent labels
Truecolor 32 channel head zebrafish Gt(desm-Citrine)ct122aR/+;Tg(kdrl:eGFP) with H2B-Cerulean, membrane-mCherry, Xanthophores, auto-fluorescence at 458nm and auto-fluorescence at 561nm. (MP4 5175 kb)
Truecolor rendering of raw data whole zebrafish with 4 fluorescent and 3 auto-fluorescent labels
Truecolor 32 channel whole zebrafish Gt(desm-Citrine)ct122aR/+;Tg(kdrl:eGFP) with H2B-Cerulean, membrane-mCherry, Xanthophores, auto-fluorescence at 458nm and auto-fluorescence at 561nm. (MP4 4840 kb)
Rendering of HySP processed 4D zebrafish head
HySP resolved z-stack of 7 color zebrafish head Gt(desm-Citrine)ct122aR/+;Tg(kdrl:eGFP) with H2B-Cerulean, membrane-mCherry, Xanthophores, auto-fluorescence at 458nm and auto-fluorescence at 561nm. (MP4 7739 kb)
Rendering of HySP processed 4D whole zebrafish
HySP resolved z-stack of 7 color zebrafish head Gt(desm-Citrine)ct122aR/+;Tg(kdrl:eGFP) with H2B-Cerulean, membrane-mCherry, Xanthophores, auto-fluorescence at 458nm and auto-fluorescence at 561nm. (MP4 7400 kb)
Rendering of HySP processed 5D zebrafish volumetric timelapse
HySP resolved 5D dataset (x,y,z,t,λ) of sample 1 28hpf zebrafish developing vasculature as detected by Tg(kdrl:eGFP); Tg(ubiq: membrane-2a-h2b-tdTomato) embryos injected with rab9-yfp and rab11-mCherry mRNA, auto-fluorescence at 561nm and 950nm. Video shows 4 views each with one channel. Dataset shape is 512x512x(25-40)x8x25 (x,y,z,λ,t). (MP4 4406 kb)
Rendering of HySP processed 5D zebrafish volumetric timelapse with 2x1 tiling
HySP resolved 5D dataset (x,y,z,t,λ) of sample 2 30hpf zebrafish developing vasculature of Tg(kdrl:eGFP) with H2B-tdTomato,membrane-Cerulean, Rab9-YFP and Rab11-mCherry, auto-fluorescence at 561nm and 950nm. Video shows 4 views each with one channel. Dataset shape is 1024x512x(15-23)x8x25 (x,y,z,λ,t). (MP4 8409 kb)
Rendering of HySP processed 5D zebrafish volumetric timelapse with 3x1 tiling
HySP resolved 5D dataset (x,y,z,t,λ) of sample 3 32hpf zebrafish developing vasculature of Tg(kdrl:eGFP) with H2B-tdTomato,membrane-Cerulean, Rab9-YFP and Rab11-mCherry, auto-fluorescence at 561nm and 950nm. Video shows 4 views each with one channel. (MP4 6442 kb)
Overlapped Rendering of HySP processed 5D zebrafish volumetric timelapse
HySP resolved 5D dataset (x,y,z,t,λ) of sample 1 28hpf zebrafish developing vasculature of Tg(kdrl:eGFP) with H2B-tdTomato,membrane-Cerulean, Rab9-YFP and Rab11-mCherry, auto-fluorescence at 561nm and 950nm. Video shows all channels together. (MP4 3671 kb)
Rendering result of multiple unmixing strategies
Optical filter, Linear Unmixing, HySP resolved z-stack of Gt(desm-Citrine)ct122aR/+;Tg(kdrl:eGFP) (MP4 26495 kb)
Rights and permissions
About this article
Cite this article
Cutrale, F., Trivedi, V., Trinh, L. et al. Hyperspectral phasor analysis enables multiplexed 5D in vivo imaging. Nat Methods 14, 149–152 (2017). https://doi.org/10.1038/nmeth.4134
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4134
This article is cited by
-
More than double the fun with two-photon excitation microscopy
Communications Biology (2024)
-
HyU: Hybrid Unmixing for longitudinal in vivo imaging of low signal-to-noise fluorescence
Nature Methods (2023)
-
Video-rate hyperspectral camera based on a CMOS-compatible random array of Fabry–Pérot filters
Nature Photonics (2023)
-
Spectral phasor analysis enables multiplexed microscopy with bioluminescent probes
Nature Methods (2022)
-
Multispectral confocal 3D imaging of intact healthy and tumor tissue using mLSR-3D
Nature Protocols (2022)