Abstract
We report the development of a 3D OrbiSIMS instrument for label-free biomedical imaging. It combines the high spatial resolution of secondary ion mass spectrometry (SIMS; under 200 nm for inorganic species and under 2 μm for biomolecules) with the high mass-resolving power of an Orbitrap (>240,000 at m/z 200). This allows exogenous and endogenous metabolites to be visualized in 3D with subcellular resolution. We imaged the distribution of neurotransmitters—gamma-aminobutyric acid, dopamine and serotonin—with high spectroscopic confidence in the mouse hippocampus. We also putatively annotated and mapped the subcellular localization of 29 sulfoglycosphingolipids and 45 glycerophospholipids, and we confirmed lipid identities with tandem mass spectrometry. We demonstrated single-cell metabolomic profiling using rat alveolar macrophage cells incubated with different concentrations of the drug amiodarone, and we observed that the upregulation of phospholipid species and cholesterol is correlated with the accumulation of amiodarone.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Elowitz, M.B., Levine, A.J., Siggia, E.D. & Swain, P.S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Zenobi, R. Single-cell metabolomics: analytical and biological perspectives. Science 342, 1243259 (2013).
Rubakhin, S.S., Lanni, E.J. & Sweedler, J.V. Progress toward single cell metabolomics. Curr. Opin. Biotechnol. 24, 95–104 (2013).
Passarelli, M.K. & Ewing, A.G. Single-cell imaging mass spectrometry. Curr. Opin. Chem. Biol. 17, 854–859 (2013).
Scannell, J.W., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191–200 (2012).
Dollery, C.T. Intracellular drug concentrations. Clin. Pharmacol. Ther. 93, 263–266 (2013).
Makarov, A. Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal. Chem. 72, 1156–1162 (2000).
Hu, Q. et al. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40, 430–443 (2005).
Kompauer, M., Heiles, S. & Spengler, B. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat. Methods 14, 90–96 (2017).
Zavalin, A., Yang, J., Hayden, K., Vestal, M. & Caprioli, R.M. Tissue protein imaging at 1 μm laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal. Bioanal. Chem. 407, 2337–2342 (2015).
Fletcher, J.S. et al. A new dynamic in mass spectral imaging of single biological cells. Anal. Chem. 80, 9058–9064 (2008).
Fletcher, J.S. & Vickerman, J.C. A new SIMS paradigm for 2D and 3D molecular imaging of bio-systems. Anal. Bioanal. Chem. 396, 85–104 (2010).
Lee, J.L.S. et al. Organic depth profiling of a nanostructured delta layer reference material using large argon cluster ions. Anal. Chem. 82, 98–105 (2010).
Körsgen, M., Pelster, A., Dreisewerd, K. & Arlinghaus, H.F. 3D ToF-SIMS analysis of peptide incorporation into MALDI matrix crystals with sub-micrometer resolution. J. Am. Soc. Mass Spectrom. 27, 277–284 (2016).
Carado, A. et al. C60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal. Chem. 80, 7921–7929 (2008).
Bruinen, A.L., Fisher, G.L. & Heeren, R.M.A. In Imaging Mass Spectrometry: Methods and Protocols (ed. Cole, L.M.) 165–173 (Springer, 2017).
Marshall, A.G., Hendrickson, C.L. & Jackson, G.S. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom. Rev. 17, 1–35 (1998).
Maharrey, S. et al. High mass resolution SIMS. Appl. Surf. Sci. 231–232, 972–975 (2004).
Smith, D.F., Robinson, E.W., Tolmachev, A.V., Heeren, R.M.A. & Paša-Tolic´ć, L. C60 secondary ion Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 83, 9552–9556 (2011).
Scheltema, R.A. et al. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics 13, 3698–3708 (2014).
Michalski, A. et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111.011015 (2011).
Seah, M.P., Havelund, R. & Gilmore, I.S. Universal equation for argon cluster size-dependence of secondary ion spectra in SIMS of organic materials. J. Phys. Chem. C 118, 12862–12872 (2014).
Passarelli, M.K. et al. Single-cell analysis: visualizing pharmaceutical and metabolite uptake in cells with label-free 3D mass spectrometry imaging. Anal. Chem. 87, 6696–6702 (2015).
Andersen, P. et al. (eds.) The Hippocampus Book (Oxford University Press, 2006).
Burgess, N., Maguire, E.A. & O'Keefe, J. The human hippocampus and spatial and episodic memory. Neuron 35, 625–641 (2002).
O'Keefe, J. & Dostrovsky, J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971).
Jones, A.R., Overly, C.C. & Sunkin, S.M. The Allen Brain Atlas: 5 years and beyond. Nat. Rev. Neurosci. 10, 821–828 (2009).
Fletcher, J.S., Rabbani, S., Henderson, A., Lockyer, N.P. & Vickerman, J.C. Three-dimensional mass spectral imaging of HeLa-M cells—sample preparation, data interpretation and visualisation. Rapid Commun. Mass Spectrom. 25, 925–932 (2011).
Jeon, S.-B., Yoon, H.J., Park, S.-H., Kim, I.-H. & Park, E.J. Sulfatide, a major lipid component of myelin sheath, activates inflammatory responses as an endogenous stimulator in brain-resident immune cells. J. Immunol. 181, 8077–8087 (2008).
Yanovsky, Y., Sergeeva, O.A., Freund, T.F. & Haas, H.L. Activation of interneurons at the stratum oriens/alveus border suppresses excitatory transmission to apical dendrites in the CA1 area of the mouse hippocampus. Neuroscience 77, 87–96 (1997).
Santarelli, L. et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 301, 805–809 (2003).
Kramar, C.P., Chefer, V.I., Wise, R.A., Medina, J.H. & Barbano, M.F. Dopamine in the dorsal hippocampus impairs the late consolidation of cocaine-associated memory. Neuropsychopharmacology 39, 1645–1653 (2014).
Papiris, S.A., Triantafillidou, C., Kolilekas, L., Markoulaki, D. & Manali, E.D. Amiodarone: review of pulmonary effects and toxicity. Drug Saf. 33, 539–558 (2010).
Anderson, N. & Borlak, J. Drug-induced phospholipidosis. FEBS Lett. 580, 5533–5540 (2006).
Mesens, N. et al. Phospholipidosis in rats treated with amiodarone: serum biochemistry and whole genome micro-array analysis supporting the lipid traffic jam hypothesis and the subsequent rise of the biomarker BMP. Toxicol. Pathol. 40, 491–503 (2012).
Shayman, J.A. & Abe, A. Drug induced phospholipidosis: an acquired lysosomal storage disorder. Biochim. Biophys. Acta 1831, 602–611 (2013).
Abe, A. & Shayman, J.A. The role of negatively charged lipids in lysosomal phospholipase A2 function. J. Lipid Res. 50, 2027–2035 (2009).
Jiang, H. et al. High-resolution sub-cellular imaging by correlative NanoSIMS and electron microscopy of amiodarone internalisation by lung macrophages as evidence for drug-induced phospholipidosis. Chem. Commun. (Camb.) 53, 1506–1509 (2017).
Lakhdar, A.A., Farish, E., Hillis, W.S. & Dunn, F.G. Long-term amiodarone therapy raises serum cholesterol. Eur. J. Clin. Pharmacol. 40, 477–480 (1991).
Zhang, D.S. et al. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature 481, 520–524 (2012).
Kraft, M.L., Weber, P.K., Longo, M.L., Hutcheon, I.D. & Boxer, S.G. Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science 313, 1948–1951 (2006).
Lovrić, J. et al. Nano secondary ion mass spectrometry imaging of dopamine distribution across nanometer vesicles. ACS Nano 11, 3446–3455 (2017).
Havelund, R. et al. Label-free imaging of biomolecules in murine brain sections using the 3D OrbiSIMS. Protocol Exchange doi:https://doi.org/10.1038/protex.2017.120 (2017).
Robinson, M.A., Graham, D.J. & Castner, D.G. ToF-SIMS depth profiling of cells: z-correction, 3D imaging, and sputter rate of individual NIH/3T3 fibroblasts. Anal. Chem. 84, 4880–4885 (2012).
Race, A.M. et al. SpectralAnalysis: software for the masses. Anal. Chem. 88, 9451–9458 (2016).
Acknowledgements
The authors thank N. Harrison, R. Reid, A. Harling and M. Skingle for their support during the project and T. Heller, M. Krehl, A. Dütting, P. Hörster, K. Strupat, S. Möhring, F. Czemper, A. Venckus, S. Kanngiesser, O. Lange and A. Kühn for excellent technical support. The authors also thank M. Tiddia for AFM measurement of frozen hydrated cell heights. This work forms part of the “3D nanoSIMS” project (ISG) in the Life-science and Health programme of the National Measurement System of the UK Department of Business, Energy and Industrial strategy. This work has received funding from the 3DMetChemIT project (ISG) of the EMPIR programme cofinanced by the Participating States and from the European Union's Horizon 2020 research and innovation programme.
Author information
Authors and Affiliations
Contributions
M.K.P., A.P. and R.H. performed experiments. M.K.P. analyzed data. P.S.M., C.F.N. and A.W. prepared tissue and cell experiments and H & E pathology. F.K. designed continuous mode Bi LMIG. R.M., A.M., D.G., E.N. designed interface to hybridize ToF and Orbitrap spectrometers. A.P., M.K.P., R.M., A.M., E.N., R.H. optimized performance of 3D OrbiSIMS. H.A. developed computer interfacing and computational methods. R.M. and E.N. designed cryo sample handling. A.W., P.S.M. and C.T.D. direction of pharmaceutical studies. M.R.A., S.H. and E.N. gave technical leadership at Thermo Fisher Scientific and ION-TOF, respectively. I.S.G. original design concept and supervised the project. M.K.P. and I.S.G. wrote the paper. All authors read and commented on the paper.
Corresponding author
Ethics declarations
Competing interests
EN is a director and shareholder of ION-TOF GmbH Muenster, Germany. AP, RM, FK, and HA are employees of ION-TOF GmbH. DG, SH and AM are employees of Thermo Fisher Scientific, the corporation that produces Orbitrap mass spectrometers. CN, PM, AW and CD (at the time of this study) are employees of GlaxoSmithKline.
Integrated supplementary information
Supplementary Figure 1 Resolving crystal violet isotope fine structure with high mass resolving power of the Orbitrap.
A) Positive ion mass spectra of crystal violet ([C25H30N3]+ at m/z 372.24371) at nominal resolution settings 240,000 (blue) and 480,000 (red) mass resolving power (mode 2) normalized to the molecular ion peak. B) Mass resolving power for secondary ions in the mass spectrum with fits of the expected m−0.5 relationship. At m/z 200 the mass resolving power is approximately 253,000 (0.5 s transient) and 417,000 (1 s transients). C) The isotopic distribution of the crystal violet molecular ion. D-F) Annotated spectra for the M+1, M+2 and M+3 isotope peaks for crystal violet spanning a dynamic range of five orders of magnitude (100% abundance to 0.001 % abundance). Results presented are from a single measurement.
Supplementary Figure 2 In situ tandem MS of cholesterol.
Orbitrap tandem MS of cholesterol fragment [M-H2O+H]+ from the corpus callosum of a mouse brain section. Peaks are annotated with chemical formulae and mass deviation. The fragment ion peaks were identified by their accurate mass. Result presented is from a single measurement.
Supplementary Figure 3 Lateral resolution measurement for the focused GCIB primary ion beam (with the secondary ion extraction potential off).
A) Ion induced secondary electron image of an electroformed mesh grid over a hole obtained with the Ar3000+ primary ion beam. Red lines along the x and y axes denote the location of linescans used to measure the resolution. B and C) Representative line scans of the secondary electron intensity across the edge of the grid for the x-axis and y-axis, respectively. D) The distribution of the FWHM lateral resolution measurements with a fit to a normal distribution function (dotted line, bin size 0.1 μm). The average FWHM lateral resolution was 1.72 μm ± 0.24 μm (μ ± 1σ) (n=246, grey bars) across the x-axis and 1.04 μm ± 0.16 μm (μ ± 1σ) (n=246, red bars) across the y-axis. Results presented are from a single image.
Supplementary Figure 4 Lateral resolution measurement for the focused GCIB primary ion beam (with the secondary ion extraction potential on, as in all SIMS analyses).
A) Ion induced secondary electron image of an electroformed mesh grid over a hole obtained with the Ar3000+ primary ion beam. Red lines along the x and y axes denote the location of linescans used to measure the resolution. B and C) Representative line scans of the secondary electron intensity across the edge of the grid for the x-axis and y-axis, respectively. D) The distribution of the FWHM lateral resolution measurements with a fit to a normal distribution function (dotted line, bin size 0.05 μm). The average FWHM lateral resolution was 2.49 μm ± 0.36 μm (μ ± 1σ) (n=243, grey bars) across the x-axis and 1.84 μm ± 0.36 μm (μ ± 1σ) (n=254, red bars) across the y-axis. Results presented are from a single image.
Supplementary Figure 5 Lateral resolution measurement for the focused 20 keV Ar3000+ GCIB primary ion beam for biomolecules.
A) Overlay ion image of selected sulfatide peaks (green), phosphatidylinositol peaks (blue) and nuclear markers (red). [Sulfatide peaks: C22(OH) ([C46H88NO12SO]− at m/z 878.6035, (0.3 ppm)), C24:1 ([C48H90NO11S]− at m/z 888.6242 (0.3 ppm)) and C24:1(OH) ([C48H90NO12S]− at m/z 904.6192 (0.3 ppm)); PI peaks: PI(38:4) ([C47H82O13P]− at m/z 885.5502 (0.3 ppm)) and PI head-group ([C6H10PO8]− at m/z 241.0120 (0.6ppm)); Nuclear markers [C4N3]− at m/z 90.0095 (2.8 ppm), [CN2O2P]− at m/z 102.9702 (0.6 ppm), [C4H2N4]− at m/z 106.0285 (0.3 ppm), [C4H3N4]− at m/z 107.0207 (0.2 ppm), [C5HN4]− at m/z 117.0207 (0.2 ppm), [C5H3N4]− at m/z 119.0363 (0.3 ppm), [C5HN4O]− at m/z 133.0156 (0.1 ppm) and [C5H4N5]− at m/z 134.0472 (6 ppb). B) Polychromatic ion image of the nuclear markers with regions of interest outlined in white. C) Detail of one nucleus from B) with line scans across the x-axis and y-axis. D) as C) for the second nucleus. Average resolution determined to be 1.34 μm ± 0.24 μm (n=5).
Supplementary Figure 6 Measurement of the Bi LMIG simultaneous lateral resolution and mass resolving power.
(A) Total ion images of ZrO2 crystals (mode 7) obtained with the Bi LMIG source with insert showing detail of thin nanostructures. B) Ion image of the [ZrO]+ peak at m/z 105.8990 (0.5 ppm). C) Ion induced SE image of the ZrO2 crystal before analysis. D) Total ion images of ZrO2 crystals (mode 7) obtained with the Bi LMIG source and insert region of interest for resolution measurement outlined in white. Linescans were obtained along the y-axis of the ion image across the ZrO2 crystal edge. E) A representative line scan shows the total ion intensity across the ZrO2 interface.. F) The distribution of lateral resolution measurements with a fit a normal distribution function (red dotted line, bin size 0.01 μm). The average FWHM lateral resolution was 172 nm ± 61 nm (μ ± 1σ) (n=95). G) The [ZrO]+ peak at m/z 105.8991 (0.2 ppm) with a FWHM width of 0.3 mDa and mass resolving power of 355,000. Results presented are from a single image.
Supplementary Figure 7 3D imaging of an Irganox delta layer reference material.
A) Orbitrap MS (mode 3), 5 keV Ar2000+ sputtering and analysis, intensity depth profile of the Irganox 1010 molecular ion, [C73H107O12]− at m/z 1175.776 (solid line), and fragment ion from Irganox 3114, [C33H46N3O5]− at m/z 564.344 (dashed line). B) ToF MS (mode 9), 5 keV Ar2000+ sputtering and 30 keV Bi3+ analysis intensity depth profile, as A) with 3D reconstruction from ToF MS data and C) Dual analyser, dual beam mode (10) with 5 keV Ar2000+ sputter beam. Intensity depth profiles as A) for Orbitrap MS Ar2000+ analysis beam and ToF MS 30 keV Bi3+ beam, with 3D reconstruction from ToF MS data. Results presented are from a single measurement.
Supplementary Figure 8 20 keV Arn GCIB Orbitrap negative ion MS of reference lipid C24:1 Mono-Sulfo Galactosyl(ß) Ceramide (d18:1/24:1) (C48H90NO11S).
(A) n = 1000, (B) n = 5500 and (C) n = 10000. Inset shows detail of the molecular ion, C48H90NO11S−, revealing little fragmentation for this particular species. Results presented are from single measurements.
Supplementary Figure 9 20 keV Arn GCIB Orbitrap positive ion MS of reference lipid 1,2-dilauroyl-sn-glycero-3-phosphoethanolamine (C29H58NO8P), normalised to the C12H22O2− peak intensity.
(A) n = 1000, (B) n = 5500 and (C) n = 10000. Results presented are from single measurements.
Supplementary Figure 10 Lipid region from the negative ion image from Figure 3D in the main text (mode 7).
The spectra was summed over the entire ion image. See Supplementary Tables 2 and 3 for annotations. Result presented is from a single measurement.
Supplementary Figure 12 Comparison of resolving power of ToF MS (mode 1) and Orbitrap MS (mode 2) for intact lipids from mouse hippocampus.
(A) Negative ion mass spectra between m/z 902 – m/z 910 (red = Orbitrap MS (mode 2), black = ToF MS (mode 1)). B) Detail of spectra between m/z 904.2 – m/z 905.0. Result presented is from a single measurement.
Supplementary Figure 13 20 keV Ar3000+ GCIB Orbitrap negative ion MS/MS of reference neurotransmitters using 20 eV collision energy.
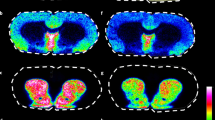
(A) GABA [M-H]−, (B) dopamine [M-H]− and (C) serotonin [M-H]−. Inset images show the co-localised spatial distribution of these secondary ions from the data in Figure 5. Results presented are from single measurements.
Supplementary Figure 14 Orbitrap MS of lipids in control and amiodarone treated macrophage cells.
The average positive ion mass spectra for control cells (n=8) and treated cells (6.25 μg/ml (n=3) and 9.38 μg/ml (n=7)).
Supplementary Figure 15 Full MS and tandem MS spectra of reference sample of amiodarone.
20 keV Ar3000+ Orbitrap MS (mode 2) spectrum (blue, positive intensity scale) and tandem MS of the [M+H]+ peak (red, negative intensity scale). Results presented are from single measurements.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Tables 1–9 (PDF 3454 kb)
Life Sciences Reporting Summary
Life Sciences Reporting Summary (PDF 160 kb)
Supplementary Protocol
Label-free Imaging of Biomolecules in Murine Brain Sections Using the 3D OrbiSIMS (PDF 682 kb)
Rights and permissions
About this article
Cite this article
Passarelli, M., Pirkl, A., Moellers, R. et al. The 3D OrbiSIMS—label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power. Nat Methods 14, 1175–1183 (2017). https://doi.org/10.1038/nmeth.4504
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4504
This article is cited by
-
Multiscale biochemical mapping of the brain through deep-learning-enhanced high-throughput mass spectrometry
Nature Methods (2024)
-
Applications of mass spectrometry imaging in botanical research
Advanced Biotechnology (2024)
-
Identifying new biomarkers of aggressive Group 3 and SHH medulloblastoma using 3D hydrogel models, single cell RNA sequencing and 3D OrbiSIMS imaging
Acta Neuropathologica Communications (2023)
-
Decoding the tumor microenvironment with spatial technologies
Nature Immunology (2023)
-
SODB facilitates comprehensive exploration of spatial omics data
Nature Methods (2023)