Abstract
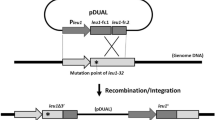
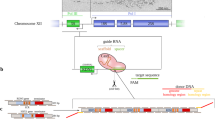
mRNA localization may be an important determinant for protein localization. We describe a simple PCR-based genomic-tagging strategy (m-TAG) that uses homologous recombination to insert binding sites for the RNA-binding MS2 coat protein (MS2-CP) between the coding region and 3′ untranslated region (UTR) of any yeast gene. Upon coexpression of MS2-CP fused with GFP, we demonstrate the localization of endogenous mRNAs (ASH1, SRO7, PEX3 and OXA1) in living yeast (Saccharomyces cerevisiae).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bashirullah, A., Cooperstock, R.L. & Lipshitz, H.D. Annu. Rev. Biochem. 67, 335–394 (1998).
Kloc, M., Zearfoss, N.R. & Etkin, L.D. Cell 108, 533–544 (2002).
Gonsalvez, G.B., Urbinati, C.R. & Long, R.M. Biol. Cell 97, 75–86 (2005).
Bertrand, E. et al. Mol. Cell 2, 437–445 (1998).
Beach, D.L., Salmon, E.D. & Bloom, K. Curr. Biol. 9, 569–578 (1999).
Abe, H. & Kool, E.T. Proc. Natl. Acad. Sci. USA 103, 263–268 (2006).
Giaever, G. et al. Nature 418, 387–391 (2002).
Ghaemmaghami, S. et al. Nature 425, 737–741 (2003).
Huh, W.K. et al. Nature 425, 686–691 (2003).
Sauer, B. Mol. Cell. Biol. 7, 2087–2096 (1987).
Aronov, S. et al. Mol. Cell. Biol.; published online 5 March 2007 (doi:10.1128/MCB01643-06).
Long, R.M. et al. Science 277, 383–387 (1997).
Takizawa, P.A., Sil, A., Swedlow, J.R., Herskowitz, I. & Vale, R.D. Nature 389, 90–93 (1997).
Aronov, S. & Gerst, J.E. J. Biol. Chem. 279, 36962–36971 (2004).
Bobola, N., Jansen, R.P., Shin, T.H. & Nasmyth, K. Cell 84, 699–709 (1996).
Mitra, D., Parnell, E.J., Landon, J.W., Yu, Y. & Stillman, D.J. Mol. Cell. Biol. 26, 4095–4110 (2006).
Grosshans, B.L. et al. J. Cell Biol. 172, 55–66 (2006).
Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P. & Tabak, H.F. Cell 122, 85–95 (2005).
Sylvestre, J., Margeot, A., Jacq, C., Dujardin, G. & Corral-Debrinski, M. Mol. Biol. Cell 14, 3848–3856 (2003).
Acknowledgements
We thank K. Bloom, P. Brennwald, J. Brickner, M. Longtine, S. Michaelis, R. Singer and D. Stillman for the generous gifts of reagents; A. Cohen and I. Goldshtein for technical assistance. This work was generously supported by a grant from the Kahn Fund for Systems Biology, Weizmann Institute of Science. J.E.G. holds the Henry Kaplan Chair in Cancer Research.
Author information
Authors and Affiliations
Contributions
Experimental design was by L.H., S.A. and J.E.G.; reagent preparation by L.H. and G.Z.; data collection, analysis, figure preparation and text by L.H. and J.E.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
PCR analysis and detection of MS2 loop integration and marker excision. (PDF 83 kb)
Supplementary Fig. 2
Sro7 is functional in SRO7::loxP::MS2L::SRO73′-UTR cells. (PDF 157 kb)
Supplementary Table 1
Oligonucleotides used in this study. (PDF 12 kb)
Supplementary Table 2
Yeast strains used in this study. (PDF 46 kb)
Rights and permissions
About this article
Cite this article
Haim, L., Zipor, G., Aronov, S. et al. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 4, 409–412 (2007). https://doi.org/10.1038/nmeth1040
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1040
This article is cited by
-
Programmable polymorphism of a virus-like particle
Communications Materials (2022)
-
Single-cell transcriptional profiling: a window into embryonic cell-type specification
Nature Reviews Molecular Cell Biology (2018)
-
P-body proteins regulate transcriptional rewiring to promote DNA replication stress resistance
Nature Communications (2017)
-
Translation in the mammalian oocyte in space and time
Cell and Tissue Research (2016)
-
Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast
Nature Communications (2015)