Abstract
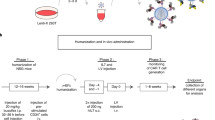
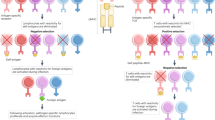
Although T-cell receptor (TCR) transgenic as well as knockout and knockin mice have had a large impact on our understanding of T-cell development, signal transduction and function, the need to cross these mice delays experiments considerably. Here we provide a methodology for the rapid expression of TCRs in mice using 2A peptide–linked multicistronic retroviral vectors to transduce stem cells of any background before adoptive transfer into RAG-1−/− mice. For simplicity, we refer to these as retrogenic mice. We demonstrate that these retrogenic mice are comparable to transgenic mice expressing three commonly used TCRs (OT-I, OT-II and AND). We also show that retrogenic mice expressing male antigen–specific TCRs (HY, MataHari and Marilyn) facilitated the analysis of positive and negative selection in female and male mice, respectively. We examined various tolerance mechanisms in epitope-coupled TCR retrogenic mice. This powerful resource could expedite the identification of proteins involved in T-cell development and function.
* Note: In the version of this article initially published, the name of one of the receptors mentioned in the abstract was incorrectly stated as OY-II instead of OT-II. The correct sentence is: “We demonstrate that these retrogenic mice are comparable to transgenic mice expressing three commonly used TCRs (OT-I, OT-II, and AND). “ This error has been corrected in the HTML and PDF versions of the article.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
01 March 2006
In the version of this article initially published, the name of one of the receptors mentioned in the abstract was incorrectly stated as OY-II instead of OT-II. The correct sentence is: “We demonstrate that these retrogenic mice are comparable to transgenic mice expressing three commonly used TCRs (OT-I, OT-II, and AND). “ This error has been corrected in the HTML and PDF versions of the article.
References
Weiss, A. T cell antigen receptor signal transduction: a tale of tails and cytoplasmic protein–tyrosine kinases. Cell 73, 209–212 (1993).
Call, M.E. & Wucherpfennig, K.W. The T-cell receptor: critical role of the membrane environment in receptor assembly and function. Annu. Rev. Immunol. 23, 101–125 (2005).
Mondino, A., Khoruts, A. & Jenkins, M.K. The anatomy of T-cell activation and tolerance. Proc. Natl. Acad. Sci. USA 93, 2245–2252 (1996).
Sommers, C.L. et al. Function of CD3 epsilon–mediated signals in T-cell development. J. Exp. Med. 192, 913–919 (2000).
Azzam, H.S. et al. Fine tuning of TCR signaling by CD5. J. Immunol. 166, 5464–5472 (2001).
Yang, L., Qin, X.F., Baltimore, D. & Van, P.L. Generation of functional antigen-specific T cells in defined genetic backgrounds by retrovirus-mediated expression of TCR cDNAs in hematopoietic precursor cells. Proc. Natl. Acad. Sci. USA 99, 6204–6209 (2002).
Yang, L. & Baltimore, D. Long–term in vivo provision of antigen-specific T-cell immunity by programming hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 102, 4518–4523 (2005).
Arnold, P.Y., Burton, A.R. & Vignali, D.A. Diabetes incidence is unaltered in glutamate decarboxylase 65–specific TCR retrogenic nonobese diabetic mice: generation by retroviral–mediated stem cell gene transfer. J. Immunol. 173, 3103–3111 (2004).
Szymczak, A.L. et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide–based retroviral vector. Nat. Biotechnol. 22, 589–594 (2004).
Donnelly, M.L. et al. The 'cleavage' activities of foot-and-mouth disease virus 2A site–directed mutants and naturally occurring '2A-like' sequences. J. Gen. Virol. 82, 1027–1041 (2001).
Donnelly, M.L. et al. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J. Gen. Virol. 82, 1013–1025 (2001).
Hogquist, K.A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
Barnden, M.J., Allison, J., Heath, W.R. & Carbone, F.R. Defective TCR expression in transgenic mice constructed using cDNA–based alpha– and beta–chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76, 34–40 (1998).
Szymczak, A.L. et al. The CD3epsilon proline-rich sequence, and its interaction with Nck, is not required for T cell development and function. J. Immunol. 175, 270–275 (2005).
Kaye, J. et al. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature 341, 746–749 (1989).
Grubin, C.E., Kovats, S., deRoos, P. & Rudensky, A.Y. Deficient positive selection of CD4 T cells in mice displaying altered repertoires of MHC class II–bound self-peptides. Immunity 7, 197–208 (1997).
Kisielow, P., Bluthmann, H., Staerz, U.D., Steinmetz, M. & von Boehmer, H. Tolerance in T cell–receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature 333, 742–746 (1988).
Lantz, O., Grandjean, I., Matzinger, P. & Di Santoz, J.P. Gamma chain required for naive CD4+ T cell survival but not for antigen proliferation. Nat. Immunol. 1, 54–58 (2000).
Valujskikh, A., Lantz, O., Celli, S., Matzinger, P. & Heeger, P.S. Cross-primed CD8(+) T cells mediate graft rejection via a distinct effector pathway. Nat. Immunol. 3, 844–851 (2002).
Hogquist, K.A., Jameson, S.C. & Bevan, M.J. Strong agonist ligands for the T cell receptor do not mediate positive selection of functional CD8+ T cells. Immunity 3, 79–86 (1995).
Ashton–Rickardt, P.G. et al. Evidence for a differential avidity model of T cell selection in the thymus. Cell 76, 651–663 (1994).
Saveanu, L., Fruci, D. & van Endert, P. Beyond the proteasome: trimming, degradation and generation of MHC class I ligands by auxiliary proteases. Mol. Immunol. 39, 203–215 (2002).
Santori, F.R. et al. Rare, structurally homologous self–peptides promote thymocyte positive selection. Immunity 17, 131–142 (2002).
Alam, S.M. et al. T cell–receptor affinity and thymocyte positive selection. Nature 381, 616–620 (1996).
Gascoigne, N.R., Zal, T. & Alam, S.M. T cell–receptor binding kinetics in T-cell development and activation. Exp. Rev. Mol. Med. 3, 1–17 (2001).
Starr, T.K., Jameson, S.C. & Hogquist, K.A. Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 (2003).
Laufer, T.M., DeKoning, J., Markowitz, J.S., Lo, D. & Glimcher, L.H. Unopposed positive selection and autoreactivity in mice expressing class II MHC only on thymic cortex. Nature 383, 81–85 (1996).
Li, W. et al. An alternate pathway for CD4 T cell development: thymocyte–expressed MHC class II selects a distinct T cell population. Immunity 23, 375–386 (2005).
Choi, E.Y. et al. Thymocyte–thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity 23, 387–396 (2005).
Jameson, S.C., Hogquist, K.A. & Bevan, M.J. Specificity and flexibility in thymic selection. Nature 369, 750–752 (1994).
Vignali, D.A. & Vignali, K.M. Profound enhancement of T cell activation mediated by the interaction between the TCR and the D3 domain of CD4. J. Immunol. 162, 1431–1439 (1999).
Acknowledgements
We are indebted to P. Matzinger, A. Rudensky, F. Carbone, M. Rubenstein, E. Parganas, S. Gingras and J. Ihle for reagents and information. We are grateful to S. Dilioglou and E. Vincent for technical assistance, K. Forbes for mouse colony management, R. Cross, J. Hoffrage and J. Smith for FACS analysis and sorting, S. Rowe for cytokine analysis, staff of the Flow Cytometry Shared Resource facility for MACS and the staff in the Hartwell Center for oligonucleotide synthesis and DNA sequencing. This work was supported by the NIH (AI–52199), a Cancer Center Support CORE grant (CA–21765) and the American Lebanese Syrian Associated Charities (ALSAC) (all to D.A.A.V.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Comparison between transgenic and retrogenic mice. (PDF 135 kb)
Supplementary Fig. 2
Characterization of AND retrogenic mice. (PDF 78 kb)
Supplementary Fig. 3
Characterization of TEa retrogenic mice. (PDF 121 kb)
Supplementary Fig. 4
Characterization of male-specific TCR retrogenic mice. (PDF 120 kb)
Supplementary Fig. 5
Characterization of epitope-coupled OT-I retrogenic mice. (PDF 160 kb)
Supplementary Table 1
Description of the TCR constructs used in this study. (PDF 76 kb)
Rights and permissions
About this article
Cite this article
Holst, J., Vignali, K., Burton, A. et al. Rapid analysis of T-cell selection in vivo using T cell–receptor retrogenic mice. Nat Methods 3, 191–197 (2006). https://doi.org/10.1038/nmeth858
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth858
This article is cited by
-
Pre-processing visualization of hyperspectral fluorescent data with Spectrally Encoded Enhanced Representations
Nature Communications (2020)
-
Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection
Nature Immunology (2020)
-
A single-domain bispecific antibody targeting CD1d and the NKT T-cell receptor induces a potent antitumor response
Nature Cancer (2020)
-
T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids
Nature Immunology (2018)
-
Generation of higher affinity T cell receptors by antigen-driven differentiation of progenitor T cells in vitro
Nature Biotechnology (2017)