Abstract
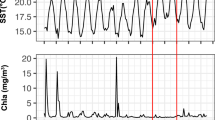
Marine phytoplankton perform approximately half of global carbon fixation, with their blooms contributing disproportionately to carbon sequestration1, and most phytoplankton production is ultimately consumed by heterotrophic prokaryotes2. Therefore, phytoplankton and heterotrophic community dynamics are important in modelling carbon cycling and the impacts of global change3. In a typical bloom, diatoms dominate initially, transitioning over several weeks to smaller and motile phytoplankton4. Here, we show unexpected, rapid community variation from daily rRNA analysis of phytoplankton and prokaryotic community members following a bloom off southern California. Analysis of phytoplankton chloroplast 16S rRNA demonstrated ten different dominant phytoplankton over 18 days alone, including four taxa with animal toxin-producing strains. The dominant diatoms, flagellates and picophytoplankton varied dramatically in carbon export potential. Dominant prokaryotes also varied rapidly. Euryarchaea briefly became the most abundant organism, peaking over a few days to account for about 40% of prokaryotes. Phytoplankton and prokaryotic communities correlated better with each other than with environmental parameters. Extending beyond the traditional view of blooms being controlled primarily by physics and inorganic nutrients, these dynamics imply highly heterogeneous, continually changing conditions over time and/or space and suggest that interactions among microorganisms are critical in controlling plankton diversity, dynamics and fates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ducklow, H. W., Steinberg, D. K. & Buesseler, K. O. Upper ocean carbon export and the biological pump. Oceanography 14, 50–58 (2001).
Kirchman, D. L. Processes in Microbial Ecology (Oxford Univ. Press, 2012).
Hutchins, D. A., Mulholland, M. R. & Fu, F. Nutrient cycles and marine microbes in a CO2-enriched ocean. Oceanography 22, 128–145 (2009).
Miller, C. B. Biological Oceanography (Blackwell, 2004).
Amin, S. A., Parker, M. S. & Armbrust, E. V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012).
Amin, S. A. et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015).
Buchan, A., LeCleir, G. R., Gulvik, C. A. & González, J. M. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nature Rev. Microbiol. 12, 686–698 (2014).
Ducklow, H. W., Kirchman, D. L., Quinby, H. L., Carlson, C. A. & Dam, H. G. Stocks and dynamics of bacterioplankton carbon during the spring bloom in the eastern North Atlantic Ocean. Deep-Sea. Res. II 40, 245–263 (1993).
Mayali, X., Palenik, B. & Burton, R. S. Dynamics of marine bacterial and phytoplankton populations using multiplex liquid bead. Environ. Microbiol. 12, 975–989 (2010).
Lindh, M. V. et al. Disentangling seasonal bacterioplankton population dynamics by high-frequency sampling. Environ. Microbiol. 17, 2459–2476 (2015).
Teeling, H. et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611 (2012).
Orsi, W. D. et al. Ecophysiology of uncultivated marine euryarchaea is linked to particulate organic matter. ISME J. 9, 1747–1763 (2015).
Mincer, T. J. et al. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9, 1162–1175 (2007).
Parada, A., Needham, D. M. & Fuhrman, J. A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time-series and global field samples. Environ. Microbiol. http://dx.doi.org/10.1111/1462-2920.13023 (2015).
Decelle, J. et al. PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol. Ecol. Resour. 15, 1435–1445 (2015).
Mann, D. G. in Cytology, Genetics and Molecular Biology of Algae (eds Chaudhary, B. R. & Agrawal, S. B. ) 249–274 (SPB Academic, 1996).
Zhu, F., Massana, R., Not, F., Marie, D. & Vaulot, D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52, 79–92 (2005).
Koumandou, V. L., Nisbet, R. E. R., Barbrook, A. C. & Howe, C. J. Dinoflagellate chloroplasts—where have all the genes gone? Trends Genet. 20, 261–267 (2004).
De Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015).
Rossini, G. P. Toxins and Biologically Active Compounds from Microalgae: Biological Effects and Risk Management Vol. 2 (CRC Press, 2014).
Schoemann, V., Becquevort, S., Stefels, J., Rousseau, V. & Lancelot, C. Phaeocystis blooms in the global ocean and their controlling mechanisms: a review. J. Sea Res. 53, 43–66 (2005).
Bienfang, P. K. Sinking rates of heterogeneous, temperate phytoplankton populations. J. Plankton Res. 3, 235–253 (1981).
Granéli, E., Salomon, P. S. & Fistarol, G. O. in Algal Toxins: Nature, Occurrence, Effect and Detection 159–178 (Springer, 2008).
Klaas, C. & Archer, D. E. Association of sinking organic matter with various types of mineral ballast in the deep sea: implications for the rain ratio. Glob. Biogeochem. Cycles 16, 1–14 (2002).
Fuhrman, J. A., Cram, J. A. & Needham, D. M. Marine microbial community dynamics and their ecological interpretation. Nature Rev. Microbiol. 13, 133–146 (2015).
Lie, A. A. Y., Kim, D. Y., Schnetzer, A. & Caron, D. A. Small-scale temporal and spatial variations in protistan community composition at the San Pedro Ocean time-series station off the coast of southern California. Aquat. Microb. Ecol. 70, 93–110 (2013).
Thompson, A. W. et al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550 (2012).
Worden, A. Z. et al. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 1257594 (2015).
Lima-Mendez, G. et al. Ocean plankton. Determinants of community structure in the global plankton interactome. Science 348, 1262073 (2015).
Vergin, K. L. et al. High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences. ISME J. 7, 1322–1332 (2013).
Noble, R. T. & Fuhrman, J. A. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14, 113–118 (1998).
Needham, D. M. et al. Short-term observations of marine bacterial and viral communities: patterns, connections and resilience. ISME J. 7, 1274–1285 (2013).
Fuhrman, J. A., Comeau, D. E., Hagstrom, A. & Chan, A. M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54, 1426–1429 (1988).
Countway, P. D., Gast, R. J., Savai, P. & Caron, D. A. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J. Eukaryot. Microbiol. 52, 95–106 (2005).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods 10, 996–998 (2013).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336 (2010).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Jeraldo, P. et al. IM-TORNADO: a tool for comparison of 16S reads from paired-end libraries. PLoS ONE 9, e114804 (2014).
Guillou, L. et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013).
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Oksanen, A. J. et al. vegan: Community Ecology Package (2015); http://vegan.r-forge.r-project.org/.
Xia, L. C. et al. Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst. Biol. 5, S15 (2011).
Xia, L. C., Ai, D., Cram, J. A., Fuhrman, J. A. & Sun, F. Efficient statistical significance approximation for local similarity analysis of high-throughput time series data. Bioinformatics 29, 230–237 (2013).
Storey, J. D. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 (2003).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
The authors acknowledge the USC Wrigley Institute of Environmental Science and thank R. Marinelli, S. Conner, G. Boivin and the crew of the Miss Christie for sampling opportunities and laboratory space, and J. Chang, C. Chow, A. Lie, S. McCallister and E. Teel for sampling assistance. The authors thank D. Caron, F. Corsetti, J. Heidelberg, E. Webb, N. Ahlgren, L. Berdjeb, J. Cram, D. Comeau, L. Gómez Consarnau, E. Fichot, M. Lee, A. Parada, B. Phillips, G. Ramirez and E. Sieradzki for discussion and feedback on the manuscript, along with R. Sachdeva who also provided computer support. This work was supported by NSF grants 1031743 and 1136818, grant GBMF3779 from the Gordon and Betty Moore Foundation Marine Microbiology Initiative and a National Science Foundation Graduate Student Research Fellowship to D.M.N.
Author information
Authors and Affiliations
Contributions
D.M.N. and J.A.F. designed the study, performed the research, analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–12, Table 1, References and Definitions. (PDF 3783 kb)
Supplementary Data Set 1
Full OTU IDs, taxonomic descriptions and abbreviations. (XLSX 2763 kb)
Rights and permissions
About this article
Cite this article
Needham, D., Fuhrman, J. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1, 16005 (2016). https://doi.org/10.1038/nmicrobiol.2016.5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.5
This article is cited by
-
Unravelling microalgal-bacterial interactions in aquatic ecosystems through 16S rRNA gene-based co-occurrence networks
Scientific Reports (2023)
-
First report on the bacterial community composition, diversity, and functions in Ramsar site of Central Himalayas, Nepal
Environmental Monitoring and Assessment (2023)
-
The microbiome of a bacterivorous marine choanoflagellate contains a resource-demanding obligate bacterial associate
Nature Microbiology (2022)
-
Contrasting sea ice conditions shape microbial food webs in Hudson Bay (Canadian Arctic)
ISME Communications (2022)
-
Annual phytoplankton dynamics in coastal waters from Fildes Bay, Western Antarctic Peninsula
Scientific Reports (2021)