Abstract
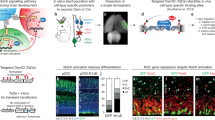
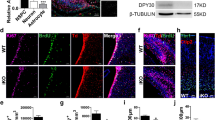
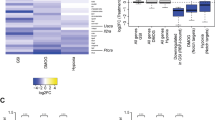
During neurogenesis, neural stem/progenitor cells (NPCs) undergo an irreversible fate transition to become neurons. The Notch pathway is important for this process, and repression of Notch-dependent Hes genes is essential for triggering differentiation. However, Notch signaling often remains active throughout neuronal differentiation, implying a change in the transcriptional responsiveness to Notch during the neurogenic transition. We identified Bcl6, an oncogene, as encoding a proneurogenic factor that is required for proper neurogenesis of the mouse cerebral cortex. BCL6 promoted the neurogenic conversion by switching the composition of Notch-dependent transcriptional complexes at the Hes5 promoter. BCL6 triggered exclusion of the co-activator Mastermind-like 1 and recruitment of the NAD+-dependent deacetylase Sirt1, which was required for BCL6-dependent neurogenesis. The resulting epigenetic silencing of Hes5 led to neuronal differentiation despite active Notch signaling. Our findings suggest a role for BCL6 in neurogenesis and uncover Notch-BCL6-Sirt1 interactions that may affect other aspects of physiology and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hsieh, J. & Gage, F.H. Chromatin remodeling in neural development and plasticity. Curr. Opin. Cell Biol. 17, 664–671 (2005).
Yoo, A.S. & Crabtree, G.R. ATP-dependent chromatin remodeling in neural development. Curr. Opin. Neurobiol. 19, 120–126 (2009).
Hirabayashi, Y. & Gotoh, Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 11, 377–388 (2010).
Bertrand, N., Castro, D.S. & Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–530 (2002).
Kageyama, R., Ohtsuka, T., Shimojo, H. & Imayoshi, I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247–1251 (2008).
Louvi, A. & Artavanis-Tsakonas, S. Notch signaling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93–102 (2006).
Pierfelice, T., Alberi, L. & Gaiano, N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840–855 (2011).
Kopan, R. & Ilagan, M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 (2009).
Ohtsuka, T., Sakamoto, M., Guillemot, F. & Kageyama, R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem. 276, 30467–30474 (2001).
Shimojo, H., Ohtsuka, T. & Kageyama, R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64 (2008).
Kawaguchi, D., Yoshimatsu, T., Hozumi, K. & Gotoh, Y. Selection of differentiating cells by different levels of delta-like 1 among neural precursor cells in the developing mouse telencephalon. Development 135, 3849–3858 (2008).
Borggrefe, T. & Liefke, R. Fine-tuning of the intracellular canonical Notch signaling pathway. Cell Cycle 11, 264–276 (2012).
Mizutani, K., Yoon, K., Dang, L., Tokunaga, A. & Gaiano, N. Differential Notch signaling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355 (2007).
Redmond, L., Oh, S.R., Hicks, C., Weinmaster, G. & Ghosh, A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat. Neurosci. 3, 30–40 (2000).
Sestan, N., Artavanis-Tsakonas, S. & Rakic, P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science 286, 741–746 (1999).
Hashimoto-Torii, K. et al. Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron 60, 273–284 (2008).
Kriegstein, A. & Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 (2009).
Noctor, S.C., Martinez-Cerdeno, V., Ivic, L. & Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004).
Miyata, T. et al. Asymmetric production of surface-dividing and non–surface-dividing cortical progenitor cells. Development 131, 3133–3145 (2004).
Malatesta, P., Hartfuss, E. & Gotz, M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127, 5253–5263 (2000).
Tiberi, L., Vanderhaeghen, P. & van den Ameele, J. Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr. Opin. Cell Biol. 24, 269–276 (2012).
Götz, M. & Huttner, W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 (2005).
Okano, H. & Temple, S. Cell types to order: temporal specification of CNS stem cells. Curr. Opin. Neurobiol. 19, 112–119 (2009).
Gaspard, N. et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455, 351–357 (2008).
Gaspard, N. & Vanderhaeghen, P. Mechanisms of neural specification from embryonic stem cells. Curr. Opin. Neurobiol. 20, 37–43 (2010).
Chang, C.C., Ye, B.H., Chaganti, R.S. & Dalla-Favera, R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA 93, 6947–6952 (1996).
Ye, B.H. et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 262, 747–750 (1993).
Iacovino, M. et al. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29, 1580–1588 (2011).
van den Ameele, J. et al. Eomesodermin induces Mesp1 expression and cardiac differentiation from embryonic stem cells in the absence of Activin. EMBO Rep. 13, 355–362 (2012).
Ye, B.H. et al. The Bcl6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16, 161–170 (1997).
Phan, R.T. & Dalla-Favera, R. The Bcl6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432, 635–639 (2004).
Otaki, J.M., Hatano, M., Matayoshi, R., Tokuhisa, T. & Yamamoto, H. The proto-oncogene Bcl6 promotes survival of olfactory sensory neurons. Dev. Neurobiol. 70, 424–435 (2010).
Zhang, S.J. et al. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53, 549–562 (2007).
Bedogni, F. et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc. Natl. Acad. Sci. USA 107, 13129–13134 (2010).
Funatsu, N., Inoue, T. & Nakamura, S. Gene expression analysis of the late embryonic mouse cerebral cortex using DNA microarray: identification of several region- and layer-specific genes. Cereb. Cortex 14, 1031–1044 (2004).
Leamey, C.A. et al. Differential gene expression between sensory neocortical areas: potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb. Cortex 18, 53–66 (2008).
Ong, C.T. et al. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 281, 5106–5119 (2006).
Sakano, D. et al. BCL6 canalizes Notch-dependent transcription, excluding Mastermind-like1 from selected target genes during left-right patterning. Dev. Cell 18, 450–462 (2010).
Herranz, D. & Serrano, M. SIRT1: recent lessons from mouse models. Nat. Rev. Cancer 10, 819–823 (2010).
Hisahara, S. et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc. Natl. Acad. Sci. USA 105, 15599–15604 (2008).
Mulligan, P. et al. A SIRT1-LSD1 co-repressor complex regulates Notch target gene expression and development. Mol. Cell 42, 689–699 (2011).
Guarani, V. et al. Acetylation-dependent regulation of endothelial Notch signaling by the SIRT1 deacetylase. Nature 473, 234–238 (2011).
Min, S.W. et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67, 953–966 (2010).
Bray, S. & Bernard, F. Notch targets and their regulation. Curr. Top. Dev. Biol. 92, 253–275 (2010).
Hoeck, J.D. et al. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat. Neurosci. 13, 1365–1372 (2010).
Sabharwal, P., Lee, C., Park, S., Rao, M. & Sockanathan, S. GDE2 regulates subtype-specific motor neuron generation through inhibition of Notch signaling. Neuron 71, 1058–1070 (2011).
Endo, K. et al. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat. Neurosci. 15, 224–233 (2012).
Prozorovski, T. et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 10, 385–394 (2008).
Gao, J. et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109 (2010).
Donmez, G., Wang, D., Cohen, D.E. & Guarente, L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142, 320–332 (2010).
Gaspard, N. et al. Generation of cortical neurons from mouse embryonic stem cells. Nat. Protoc. 4, 1454–1463 (2009).
Heng, J.I. et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455, 114–118 (2008).
Rustighi, A. et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat. Cell Biol. 11, 133–142 (2009).
Takeda, N. et al. Bcl6 is a transcriptional repressor for the IL-18 gene. J. Immunol. 171, 426–431 (2003).
Saito, T. & Nakatsuji, N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240, 237–246 (2001).
Dufour, A. et al. Area specificity and topography of thalamocortical projections are controlled by ephrin/Eph genes. Neuron 39, 453–465 (2003).
Depaepe, V. et al. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature 435, 1244–1250 (2005).
Acknowledgements
We thank G. Vassart for continuous support and interest, D. Christophe and members of the Vanderhaeghen lab and Institute for Interdisciplinary Research for helpful discussions and advice, F. Bollet-Quivogne (Fonds National de la Recherche Scientifique (FNRS) Logistic Scientist) of the Light Microscopy Facility for his support with imaging, R. Dalla-Favera (Columbia University) for generously sharing Bcl6−/− mice, F. Guillemot (National Institute of Medical Research) for kindly providing P19 cells, and G. Del Sal ((Laboratorio Nazionale CIB) Trieste) for the pCS2-NΔE construct, and B. Hassan, A. Soldano and K. De Backer for critically reading the manuscript. This work was funded by grants from the Belgian Queen Elizabeth Medical Foundation, the Fondations Pierre Clerdent and Roger de Spoelberch, the Action de Recherches Concertées Programs, the Interuniversity Attraction Poles Program, Belgian State, Federal Office for Scientific, Technical and Cultural Affairs, the Belgian FNRS and Fonds pour la Recherche Scientifique Médicale, and the Welbio and Programme d'Excellence CIBLES of the Walloon Region (to P.V.), as well as an EMBO Long-Term Fellowship (to L.T.) and a Marie Curie Fellowship (to T.B.). P.V. is Research Director, L.T. Postdoctoral Fellow, and J.v.d.A. and J.P. Research Fellows of the FNRS.
Author information
Authors and Affiliations
Contributions
L.T., J.v.d.A., J.D., J.P., D.G., A.H., A.B. and J.B. performed all experiments. T.B., M.I. and M.K. provided crucial cell reagents. L.T., J.v.d.A. and P.V. designed and analyzed all experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Table 1 (PDF 3936 kb)
Rights and permissions
About this article
Cite this article
Tiberi, L., van den Ameele, J., Dimidschstein, J. et al. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat Neurosci 15, 1627–1635 (2012). https://doi.org/10.1038/nn.3264
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3264
This article is cited by
-
SIRT1, miR-132 and miR-212 link human longevity to Alzheimer’s Disease
Scientific Reports (2018)
-
Genome-wide DNA Methylation and RNAseq Analyses Identify Aberrant Signalling Pathways in Focal Cortical Dysplasia (FCD) Type II
Scientific Reports (2018)
-
The brain, sirtuins, and ageing
Nature Reviews Neuroscience (2017)
-
Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis
Nature Communications (2016)
-
Notch signalling in context
Nature Reviews Molecular Cell Biology (2016)