Abstract
Ten-eleven translocation (TET) enzymes mediate the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which is enriched in brain, and its ultimate DNA demethylation. However, the influence of TET and 5hmC on gene transcription in brain remains elusive. We found that ten-eleven translocation protein 1 (TET1) was downregulated in mouse nucleus accumbens (NAc), a key brain reward structure, by repeated cocaine administration, which enhanced behavioral responses to cocaine. We then identified 5hmC induction in putative enhancers and coding regions of genes that have pivotal roles in drug addiction. Such induction of 5hmC, which occurred similarly following TET1 knockdown alone, correlated with increased expression of these genes as well as with their alternative splicing in response to cocaine administration. In addition, 5hmC alterations at certain loci persisted for at least 1 month after cocaine exposure. Together, these reveal a previously unknown epigenetic mechanism of cocaine action and provide new insight into how 5hmC regulates transcription in brain in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (suppl.), 245–254 (2003).
Weaver, I.C. et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 (2004).
Miller, C.A. & Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 53, 857–869 (2007).
Ma, D.K. et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009).
Feng, J. et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430 (2010).
Ooi, S.K. & Bestor, T.H. The colorful history of active DNA demethylation. Cell 133, 1145–1148 (2008).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
Ko, M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010).
He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Wu, H. & Zhang, Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25, 2436–2452 (2011).
Branco, M.R., Ficz, G. & Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 (2012).
Pastor, W.A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Guo, J.U., Su, Y., Zhong, C., Ming, G.L. & Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011).
Globisch, D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5, e15367 (2010).
Szwagierczak, A., Bultmann, S., Schmidt, C.S., Spada, F. & Leonhardt, H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 38, e181 (2010).
Szulwach, K.E. et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616 (2011).
Rudenko, A. et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122 (2013).
Kaas, G.A. et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA Demethylation, gene transcription and memory formation. Neuron 79, 1086–1093 (2013).
Zhang, R.R. et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245 (2013).
Li, X. et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc. Natl. Acad. Sci. USA 111, 7120–7125 (2014).
Nestler, E.J. Molecular basis of long-term plasticity underlying addiction. Nat. Rev. Neurosci. 2, 119–128 (2001).
Feng, J. & Nestler, E.J. Epigenetic mechanisms of drug addiction. Curr. Opin. Neurobiol. 23, 521–528 (2013).
LaPlant, Q. et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 13, 1137–1143 (2010).
Le, T., Kim, K.P., Fan, G. & Faull, K.F. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Anal. Biochem. 412, 203–209 (2011).
Rajagopal, N. et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLOS Comput. Biol. 9, e1002968 (2013).
Malik, A.N. et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat. Neurosci. 17, 1330–1339 (2014).
Heintzman, N.D. et al. Histone modifications at human enhancers reflect global cell type–specific gene expression. Nature 459, 108–112 (2009).
Creyghton, M.P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Feng, J. et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 15, R65 (2014).
Sasai, N., Nakao, M. & Defossez, P.A. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 38, 5015–5022 (2010).
Huh, G.S. et al. Functional requirement for class I MHC in CNS development and plasticity. Science 290, 2155–2159 (2000).
Vogel-Ciernia, A. et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 16, 552–561 (2013).
Booth, M.J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Zachariou, V. et al. Distinct roles of adenylyl cyclases 1 and 8 in opiate dependence: behavioral, electrophysiological and molecular studies. Biol. Psychiatry 63, 1013–1021 (2008).
Graham, D.L. et al. Tropomyosin-related kinase B in the mesolimbic dopamine system: region-specific effects on cocaine reward. Biol. Psychiatry 65, 696–701 (2009).
Lobo, M.K. et al. Cell type–specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science 330, 385–390 (2010).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Khare, T. et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 19, 1037–1043 (2012).
Mitchell, A.C. et al. The genome in three dimensions: a new frontier in human brain research. Biol. Psychiatry 75, 961–969 (2014).
Kim, T.K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Natoli, G. & Andrau, J.C. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 46, 1–19 (2012).
Pennacchio, L.A. et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 (2006).
Cruz, F.C. et al. New technologies for examining the role of neuronal ensembles in drug addiction and fear. Nat. Rev. Neurosci. 14, 743–754 (2013).
Mellén, M., Ayata, P., Dewell, S., Kriaucionis, S. & Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012).
Hellman, A. & Chess, A. Gene body–specific methylation on the active X chromosome. Science 315, 1141–1143 (2007).
Wu, H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 (2011).
Robison, A.J. et al. Behavioral and structural responses to chronic cocaine require a feedforward loop involving DeltaFosB and calcium/calmodulin-dependent protein kinase II in the nucleus accumbens shell. J. Neurosci. 33, 4295–4307 (2013).
Vialou, V. et al. DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat. Neurosci. 13, 745–752 (2010).
Li, N. et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 329, 959–964 (2010).
Song, C.X. et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 29, 68–72 (2011).
Vialou, V. et al. Serum response factor and cAMP response element binding protein are both required for cocaine induction of DeltaFosB. J. Neurosci. 32, 7577–7584 (2012).
Booth, M.J. et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 8, 1841–1851 (2013).
Krueger, F. & Andrews, S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Shen, L. et al. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE 8, e65598 (2013).
Ernst, J. & Kellis, M. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012).
Shen, L., Shao, N., Liu, X. & Nestler, E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284 (2014).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Bailey, T.L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Huang, D.W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Acknowledgements
We thank A. Chess for critical comments and O. Jabado and M. Mahajan from the Mount Sinai Genomics Core for technical support. This work was supported by grants from the National Institute on Drug Abuse (E.J.N.), the National Institutes of Health (P.J., P.C., G.F., K.F.F. and H.S.) and the Simons Foundation (H.S.).
Author information
Authors and Affiliations
Contributions
The studies were conceived and designed by E.J.N., P.J. and J.F. J.F. performed RNA-seq and ChIP-seq. K.E.S., Y.L. and J.F. performed 5hmC capture and sequencing. L.S., N.S. and J.F. performed bioinformatic analyses. J.F. and J.H. performed oxBS-seq. JF., V.V., B.M.L., V.S. and I.M. performed qPCR analyses. V.V., D.E., T.C. and J.F. performed immunohistochemistry. J.F., V.V., D.F., J.K. and E.R. performed stereotaxic surgeries and behavioral assays. T.L., K.F.F. and G.F. contributed LC-ESI-MS/MS data. C.Z., G.M. and H.S. provided AAV-Tet1 shRNA and AAV-TET1 viruses. M.E.C. performed western blotting. P.C. contributed reagents. G.T. contributed human samples. D.F., B.L., B.M.L., V.S. and P.K. helped to prepare the samples and collect the data. The paper was written by J.F. and E.J.N. and was edited by the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Representative immunohistochemistry images and TET western blots.
a, TET1 staining in NAc of mice treated with repeated cocaine or saline. A decrease in TET1 levels after cocaine is seen. b, Full length western blots for TET1, TET2, TET3, and β-actin. Saline (s) and cocaine (c) samples were loaded alternatively. The blue rectangles demonstrate the lanes cropped in Figure 1c. c, DAPI/GFP/DARPP-32 staining of a mouse brain section after viral injection. DAPI staining demonstrates intact brain structure of the injected hemisphere. GFP staining illustrates the spread of viral-mediated transgene expression in NAc. DARPP-32 immunohistochemistry indicates intact striatum structure after viral injection.
Supplementary Figure 2 Validation of virally mediated Tet1 shRNA knockdown and overexpression.
a, qPCR indicates Tet1 transcript levels are decreased after Tet1 viral knockdown (KD) in NAc. Student unpaired t-test P=0.0026, t(18)=3.494. N=10 per group. b, Quantification by Western blotting confirms TET1 protein levels are also decreased after Tet1 viral knockdown in this brain region. Student unpaired t-test, P=0.047, t(12)=1.805. N=8 per group. Box plots present, in ascending order, minimum sample value, first quartile, median, third quartile, maximum sample value. * indicates P<0.05, ** indicates P<0.01. c, DNA gel electrophoresis picture demonstrates the specific amplification of human TET1 that is encoded by the viral overexpression (OE) construct in mouse NAc.
Supplementary Figure 3 Relative expression of Tet mRNAs in adult mouse NAc.
qPCR analysis demonstrates relative abundance of Tet1, Tet2, and Tet3 transcripts in adult male mouse NAc. N=9 per group. Data are presented as mean ± SEM.
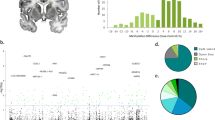
Supplementary Figure 4 Distribution of 5hmC-enriched regions in NAc of saline- and cocaine-treated mice.
a, Histogram of 5hmC peak counts (in numbers per million bp sequencing reads) of each chromosome in saline and repeated cocaine samples. b, Circular layout of 5hmC peaks under saline conditions in blue bars below the black-white ideogram along chromosomal coordinates. 5hmC differential sites after repeated cocaine are shown in yellow (red/green) bars of the inside circle. c, Coverage plot of 5hmC peak regions under saline (orange color) and cocaine (green color) conditions from transcription start sites (TSSs) to transcription ending sites (TESs), as well as up- and downstream regions, of genes genome-wide. The coverage of up- and downstream interpolated 33% sequence is also shown. DNA input trace is in purple color.
Supplementary Figure 5 Selective enrichment of H3K4me1, H3K27ac and 5hmC at enhancer regions.
ChIP-seq for H3K4me1 and H3K27ac as well as 5hmC-seq are plotted over the mouse brain enhancer regions defined by the ENCODE project (http://genome.ucsc.edu/ENCODE/). Coverage plot of H3K4me1 (blue color), H3K27ac (orange color), and 5hmC (green color) from 1500 bp up- and downstream of enhancer sites reveals their enrichment at enhancer regions as demonstrated by sequencing read counts while compared to input which has similar read counts as H3K4me3 (data not shown).
Supplementary Figure 6 ChIP-qPCR validation of putative enhancers.
a, H3K4me1 ChIP followed by qPCR from a separate cohort of animals display significant enrichments of H3K4me1 at 10 putative enhancer sites as predicted by H3K4me1 and H3K27ac ChIP-seq co-binding. Student’s unpaired t-test for enhancers1 to 10: P=0.00008, t(14)=5.455; P=0.00001, t(14)=6.832; P=0.00143, t(14)=3.958; P=0.00005, t(14)=5.713; P=0.00001, t(14)=6.527; P=0.00005, t(14)=5.717; P=0.00013, t(14)=5.213; P=0.04619, t(14)=2.187; P=0.00050, t(14)=4.495; P=0.00004, t(14)=5.842. N=8/group. b, similar validation is observed for H3K27ac ChIP enrichment on the same putative enhancer sites. Student’s unpaired t-test for enhancers1 to 10: P=0.00008, t(14)=5.507; P=0.00008, t(14)=5.481; P=0.00255, t(14)=3.665; P=2.94444E-08, t(14)=10.97; P=3.7615831E-09, t(14)=12.88; P=2.9862057E-08, t(14)=10.95; P=5.1980284E-04, t(14)=4.479; P=0.00624, t(14)=3.214; P=0.00042, t(14)=4.586; P=0.00007, t(14)=5.566. N=8/group. c, P300 ChIP demonstrates an enrichment at most of the 10 putative enhancer sites. Student’s unpaired t-test for enhancers1 to 10: P=0.06760, t(11)=2.01; P=0.01612, t(11)=2.839; P=0.08876, t(11)=1.881; P=0.00036, t(11)=5.071; P=0.00026, t(11)=5.241; P=0.00123, t(11)=4.312; P=0.00091, t(11)=4.477; P=0.00155, t(11)=4.177; P=0.00820, t(11)=3.212; P=0.02076, t(11)=2.748. All putative enhancer bindings are compared to a non-putative enhancer control region at OCT4 gene promoter (-400bp to -550bp). All experiments are done under saline basal codition. The 10 randomly selected putative enhancer sites are chr8:124192000-124192600, chr12:32746400-32746800, chr13:49223800-49225000, chrX:53603400-53603600, chr13:74702000-74702200, chr18:38527400-38527600, chr12:118582600-118583200, chr17:25690800-25691000, chr9:61420400-61421200, chr3:90251000-90251400. Conrol n=6/group, enhancer n=7/group. Data are presented as mean ± SEM. * indicates P<0.05, ** indicates P<0.01, *** indicates P<0.001.
Supplementary Figure 7 Gene ontology of genes that show cocaine-induced changes by both 5hmC-seq and RNA-seq.
a, Gene ontology of the most significant enriched functional annotation groups of genes that display both mRNA induction (24 hr) and gene body 5hmC induction. b, Gene ontology of the most significant enriched annotation groups of genes that display both mRNA induction (4 hr after a cocaine challenge) and gene body 5hmC induction prior to the challenge dose of cocaine.
Supplementary Figure 8 oxBS-seq methodology validation.
Two loci with a significant increase (4932411E22Rik) or decrease (Rsbn1l) of 5hmC in NAc after repeated cocaine were selected from 5hmC-seq analysis. a, oxBS-seq confirmed that Rsbn1l shows decreased 5hmC frequency at all seven CpG sites. This decrease is statistically significant across the Rsbn1l locus. Student paired t-test, P=3.12E-08, t(6)=9.501. b, oxBS-seq confirmed that 4932411E22Rik displays increased 5hmC frequency at most CpG sites tested. This increase is statistically significant across the 4932411E22Rik locus. Student paired t-test, P=0.005, t(9)=3.687. N=1-2 biological replicates/condition. Box plots present, in ascending order, minimum sample value, first quartile, median, third quartile, maximum sample value. ** indicates P<0.01, *** indicates P<0.001.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1, 5 and 7 (PDF 1343 kb)
Supplementary Table 2
Gene regions that display cocaine-induced changes in 5hmC (XLSX 595 kb)
Supplementary Table 3
Genes whose putative nearby enhancers show cocaine-induced switches to chromatin state 4 or 5 (XLS 1616 kb)
Supplementary Table 4
Gene ontology of genes whose putative nearby enhancers show cocaine-induced switches to chromatin states 4 or 5 (XLSX 22 kb)
Supplementary Table 6
Genes that show overlap in cocaine regulation of 5hmC and alternative splicing (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Feng, J., Shao, N., Szulwach, K. et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci 18, 536–544 (2015). https://doi.org/10.1038/nn.3976
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3976
This article is cited by
-
Cell-type specific transcriptional adaptations of nucleus accumbens interneurons to amphetamine
Molecular Psychiatry (2023)
-
Synaptic Mechanisms of Delay Eyeblink Classical Conditioning: AMPAR Trafficking and Gene Regulation in an In Vitro Model
Molecular Neurobiology (2023)
-
Psychostimulant-induced aberrant DNA methylation in an in vitro model of human peripheral blood mononuclear cells
Clinical Epigenetics (2022)
-
Cocaine-related DNA methylation in caudate neurons alters 3D chromatin structure of the IRXA gene cluster
Molecular Psychiatry (2021)
-
Regulation of dopamine-dependent transcription and cocaine action by Gadd45b
Neuropsychopharmacology (2021)