Abstract
Schizophrenia is a devastating psychiatric illness with high heritability. Brain structure and function differ, on average, between people with schizophrenia and healthy individuals. As common genetic associations are emerging for both schizophrenia and brain imaging phenotypes, we can now use genome-wide data to investigate genetic overlap. Here we integrated results from common variant studies of schizophrenia (33,636 cases, 43,008 controls) and volumes of several (mainly subcortical) brain structures (11,840 subjects). We did not find evidence of genetic overlap between schizophrenia risk and subcortical volume measures either at the level of common variant genetic architecture or for single genetic markers. These results provide a proof of concept (albeit based on a limited set of structural brain measures) and define a roadmap for future studies investigating the genetic covariance between structural or functional brain phenotypes and risk for psychiatric disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sullivan, P.F., Daly, M.J. & O'Donovan, M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 13, 537–551 (2012).
Purcell, S.M. et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190 (2014).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 18, 199–209 (2015).
van Erp, T.G. et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry 10.1038/mp.2015.63 (2 June 2015).
Haijma, S.V. et al. Brain volumes in schizophrenia: a meta-analysis in over 18 000 subjects. Schizophr. Bull. 39, 1129–1138 (2013).
Boos, H.B., Aleman, A., Cahn, W., Hulshoff Pol, H. & Kahn, R.S. Brain volumes in relatives of patients with schizophrenia: a meta-analysis. Arch. Gen. Psychiatry 64, 297–304 (2007).
Thermenos, H.W. et al. A review of neuroimaging studies of young relatives of individuals with schizophrenia: a developmental perspective from schizotaxia to schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 162B, 604–635 (2013).
Hibar, D.P. et al. & Alzheimer's Disease Neuroimaging Initiative; CHARGE Consortium; EPIGEN; IMAGEN; SYS. Common genetic variants influence human subcortical brain structures. Nature 520, 224–229 (2015).
Blokland, G.A., de Zubicaray, G.I., McMahon, K.L. & Wright, M.J. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res. Hum. Genet. 15, 351–371 (2012).
Stefansson, H. et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature 505, 361–366 (2014).
Bulik-Sullivan, B.K. et al. & Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Lee, S.H. et al. Cross-Disorder Group of the Psychiatric Genomics Consortium; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 45, 984–994 (2013).
Bulik-Sullivan, B. et al. & ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Purcell, S.M. et al. & International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Plaisier, S.B., Taschereau, R., Wong, J.A. & Graeber, T.G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010).
Ho, Y.Y.W. et al. Common genetic variants influence whorls in fingerprint patterns. J. Invest. Dermatol. (in the press).
Nichols, T., Brett, M., Andersson, J., Wager, T. & Poline, J.B. Valid conjunction inference with the minimum statistic. Neuroimage 25, 653–660 (2005).
Rose, E.J. & Donohoe, G. Brain vs behavior: an effect size comparison of neuroimaging and cognitive studies of genetic risk for schizophrenia. Schizophr. Bull. 39, 518–526 (2013).
Mier, D., Kirsch, P. & Meyer-Lindenberg, A. Neural substrates of pleiotropic action of genetic variation in COMT: a meta-analysis. Mol. Psychiatry 15, 918–927 (2010).
Hariri, A.R. & Weinberger, D.R. Imaging genomics. Br. Med. Bull. 65, 259–270 (2003).
Witte, J.S., Visscher, P.M. & Wray, N.R. The contribution of genetic variants to disease depends on the ruler. Nat. Rev. Genet. 15, 765–776 (2014).
Wood, A.R. et al. & Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Toulopoulou, T. et al. Reciprocal causation models of cognitive vs volumetric cerebral intermediate phenotypes for schizophrenia in a pan-European twin cohort. Mol. Psychiatry 20, 1386–1396 (2015).
Minzenberg, M.J., Laird, A.R., Thelen, S., Carter, C.S. & Glahn, D.C. Meta-analysis of 41 functional neuroimaging studies of executive function in schizophrenia. Arch. Gen. Psychiatry 66, 811–822 (2009).
Narr, K.L. et al. Mapping cortical thickness and gray matter concentration in first episode schizophrenia. Cereb. Cortex 15, 708–719 (2005).
Weinberger, D.R. On the plausibility of “the neurodevelopmental hypothesis” of schizophrenia. Neuropsychopharmacology 14 (suppl. 3), 1S–11S (1996).
Visscher, P.M., Brown, M.A., McCarthy, M.I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Ge, T. et al. Massively expedited genome-wide heritability analysis (MEGHA). Proc. Natl. Acad. Sci. USA 112, 2479–2484 (2015).
Gottesman, I.I. & Gould, T.D. The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry 160, 636–645 (2003).
Kendler, K.S. & Neale, M.C. Endophenotype: a conceptual analysis. Mol. Psychiatry 15, 789–797 (2010).
Cannon, T.D. & Keller, M.C. Endophenotypes in the genetic analyses of mental disorders. Annu. Rev. Clin. Psychol. 2, 267–290 (2006).
Fuchsberger, C., Abecasis, G.R. & Hinds, D.A. minimac2: faster genotype imputation. Bioinformatics 31, 782–784 (2015).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26, 2190–2191 (2010).
Deng, X., Xu, J. & Wang, C. Improving the power for detecting overlapping genes from multiple DNA microarray-derived gene lists. BMC Bioinformatics 9 (suppl. 6), S14 (2008).
Storey, J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 63, 479–498 (2002).
Rietveld, C.A. et al. & LifeLines Cohort Study. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471 (2013).
Acknowledgements
PGC. The authors are grateful to the many family members who participated in the studies that recruited these samples, to the many clinicians who assisted in their recruitment, and to our team members, without whom this study would have been impossible. Core funding for the PGC is from the US National Institute of Mental Health (NIMH; U01 MH094421). Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org/) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480-05-003), along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. The GRAS data collection was supported by the Max Planck Society, the Max-Planck-Förderstiftung and the DFG Center for Nanoscale Microscopy & Molecular Physiology of the Brain (CNMPB), Göttingen, Germany. The Boston CIDAR project was supported by the NIMH (P50 MH080272, R.W.M.; U01 MH081928, L.J.S.; R01 MH092380, T.L.P.) and the Massachusetts General Hospital Executive Committee on Research (T.L.P.). P.H.L. is supported by NIMH K99 MH101367. ISC Portugal: C.N.P. and M.T.P. have been supported by NIMH grants MH085548, MH085542, MH071681, MH061884, MH58693 and MH52618 and NCRR grant RR026075. C.N.P., M.T.P. and A.H.F. have been supported by grants from the Department of Veterans Affairs Merit Review Program. The Danish Aarhus study was supported by grants from Lundbeck Foundation, Danish Strategic Research Council, Aarhus University, and Stanley Research Foundation. Work in Cardiff was supported by UK Medical Research Council (MRC) Centre (G0800509) and MRC Programme (G0801418) grants, the European Community's Seventh Framework Programme (HEALTH-F2-2010-241909, Project EU-GEI) the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement 279227, a fellowship to J.W. from the MRC/Welsh Assembly Government and the Margaret Temple Award from the British Medical Association. We thank Novartis for their input in obtaining CLOZUK samples, staff at The Doctor's Laboratory (L. Levett and A. Levett) for help with sample acquisition and data linkage, and staff in Cardiff (K. Mantripragada and L. Hopkins) for sample management. CLOZUK and some other samples were genotyped at the Broad Institute or by the Wellcome Trust Case Control Consortium (WTCCC) and Wellcome Trust Case Control Consortium 2 (WTCCC2) (WT 083948/Z/07/Z). We acknowledge use of the British 1958 Birth Cohort DNA (MRC: G0000934) and the Wellcome Trust (068545/Z/0 and 076113/C/04/Z), the UK Blood Services Common Controls (UKBS-CC collection), funded by the Wellcome Trust (076113/C/04/Z) and by a National Institute for Health Research programme grant to National Health Service Blood and Transplant (RP-PG-0310-1002). Virginia Commonwealth University investigators were supported by NIMH grants R01 MH083094, R01 MH041953, and R01 MH068881 and WTCCC2 grant WTCCC-084710. Recruitment of families in Bulgaria was funded by the Janssen Research Foundation, Beerse, Belgium. We thank the staff in the Neuroscience Biomarkers Genomic Lab led by R. Favis at Janssen for sample processing and the staff at Illumina for genotyping Janssen DNA samples. We also thank A. Santos, N. Bottrel, M.-A. Franc and W. Cafferty of Janssen Research & Development) for operational support. Dutch samples were funded by the Netherlands Organization for Health Research and Development (ZonMw) in the Mental Health program and by NIMH R01 MH078075. Danish samples were funded by the Danish Council for Strategic Research (Journ.nr. 09-067048), the Danish National Advanced Technology Foundation (Journ.nr. 001-2009-2), the Lundbeck Foundation (Journ.nr. R24-A3243), and the EU 7th Framework Programme. The Wellcome Trust supported this study as part of the WTCCC2 project. E. Bramon holds a MRC New Investigator Award and a MRC Centenary Award. The TOP Study was supported by the Research Council of Norway (213837, 217776, 223273), South-East Norway Health Authority (2013-123) and K.G. Jebsen Foundation. This work was supported by the Donald and Barbara Zucker Foundation, the North Shore – Long Island Jewish Health System Foundation and grants from the Stanley Foundation (A.K.M.), NARSAD (A.K.M.), NIMH (MH065580 to T. Lencz; MH001760 to A.K.M.), and NIMH RC2 MH089964 and R01 MH084098. Finnish samples were funded by SynSys (EU FP7-242167), Sigrid Juselius Foundation, the Academy of Finland (grant 251704), and the Sohlberg Foundation. The Swedish Research Council (grants 2006-4472, 2009-5269, 2009-3413) and the County Councils of Västerbotten and Norrbotten, Sweden, supported the collection of the Umeå samples. The Betula Study, from which the Umeå controls were recruited, is supported by grants from the Swedish Research Council (grants 345-2003-3883, 315-2004-6977) and the Bank of Sweden Tercentenary Foundation, the Swedish Council for Planning and Coordination of Research, the Swedish Council for Research in the Humanities and Social Sciences and the Swedish Council for Social Research. We acknowledge support from NIMH K01 MH085812 (M.C.K.) and NIMH R01 MH100141 (M.C.K.). Estonian Genome Center at the University of Tartu (EGCUT) work was supported by Targeted Financing from the Estonian Ministry of Science and Education (SF0180142s08), US National Institutes of Health (NIH) grant R01 DK075787, the Development Fund of the University of Tartu (grant SP1GVARENG), the European Regional Development Fund to the Centre of Excellence in Genomics (EXCEGEN; grant 3.2.0304.11-0312) and FP7 grant 313010. M. Macek was supported by CZ.2.16/3.1.00/24022OPPK, NT/13770-4 and 00064203 FN Motol. Funding from the Singapore National Medical Research Council (NMRC/TCR/003/2008) and the Singapore Biomedical Research Council. We acknowledge the support of the Singapore Agency for Science, Technology and Research (A*STAR). Genotyping of the Swedish Hubin sample was performed by the SNP&SEQ Technology Platform in Uppsala, which is supported by Uppsala University, Uppsala University Hospital, Science for Life Laboratory and the Swedish Research Council (contracts 80576801 and 70374401). The Swedish Hubin sample was supported by Swedish Research Council (I.A., E.G.J.) and Stockholm County Council and the Karolinska Insititutet (E.G.J.). B.J.M., V.J.C., R.J.S., S.V.C., F.A.H., A.V.J., C.M.L., P.T.M., C.P. and U.S. were supported by the Australian Schizophrenia Research Bank, which is supported by an enabling grant from the National Health and Medical Research Council (386500), the Pratt Foundation, Ramsay Health Care, the Viertel Charitable Foundation, the Schizophrenia Research Institute and the NSW Department of Health. C.P. is supported by a Senior Principal Research Fellowship from the National Health and Medical Research Council (Australia). The Perth sample collection was funded by Australian National Health and Medical Research Council project grants and the Australian Schizophrenia Research Bank. The Bonn/Mannheim sample was genotyped in a study that was supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Genome Research Network MooDS (Systematic Investigation of the Molecular Causes of Major Mood Disorders and Schizophrenia; grant 01GS08144 to M.M.N. and S.C., grant 01GS08147 to M.R.), under the National Genome Research Network plus (NGFNplus), and the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under e:Med Programme (GlaxoSmithKline control sample; B.M.-M.) This work has been funded by the Bavarian Ministry of Commerce and by the BMBF in the framework of the National Genome Research Network, Förderkennzeichen 01GS0481 and the Bavarian Ministry of Commerce. M.M.N. is a member of the German Research Foundation (DFG)-funded Excellence Cluster ImmunoSensation. M.M.N. also received support from the Alfried Krupp von Bohlen und Halbach-Stiftung. M.R. was supported by the 7th Framework Programme of the European Union (ADAMS project, HEALTH-F4-2009-242257; CRESTAR project, HEALTH-2011-1.1-2) grant 279227. J. Knight holds the Joanne Murphy Professor in Behavioural Science. The Stanley Center for Psychiatric Research at the Broad Institute acknowledges funding from the Stanley Medical Research Institute. Support for the Sweden Schizophrenia Study (principal investigators P.F.S., C.M.H. and P. Sklar) was provided by the NIMH (R01 MH077139 and R01 MH095034), the Stanley Center for Psychiatric Research, the Sylvan Herman Foundation, the Friedman Brain Institute at the Mount Sinai School of Medicine, the Karolinska Institutet, Karolinska University Hospital, the Swedish Research Council, the Swedish County Council and the Söderström Königska Foundation. We acknowledge use of DNA from the UK Blood Services Collection of Common Controls (UKBS collection), funded by the Wellcome Trust grant 076113/CI04/Z, by Juvenile Diabetes Research Foundation grant WT0618S8, and by the National Institute of Health Research of England. The Multicenter Genetics Studies of Schizophrenia and Molecular Genetics of Schizophrenia studies study were supported by NIMH grant R01 MH062276 (to D.F.L., C.L., M.J.O. and D.B.W.), grant R01 MH068922 (to P.V.G.), grant R01 MH068921 (to A.E.P.) and grant R01 MH068881 (to B.P.R.). D.F.L. was supported by the Walter E. Nichols, M.D., Professorship in the School of Medicine, the Eleanor Nichols Endowment, the Walter F. & Rachael L. Nichols Endowment and the William and Mary McIvor Endowment, Stanford University. This study was supported by NIH R01 grants (MH67257 to N.G.B., MH59588 to B.J.M., MH59571 to P.V.G., MH59565 to R.F., MH59587 to F.A., MH60870 to W.F.B., MH59566 to D.W.B., MH59586 to J.M.S., MH61675 to D.F.L., MH60879 to C.R.C. and MH81800 to P.V.G.), NIH U01 grants (MH46276 to C.R.C., MH46289 to C. Kaufmann, MH46318 to M.T. Tsuang, MH79469 to P.V.G. and MH79470 to D.F.L.), the Genetic Association Information Network (GAIN) and The Paul Michael Donovan Charitable Foundation. Genotyping was carried out by the Center for Genotyping and Analysis at the Broad Institute of Harvard and MIT (S. Gabriel and D.B. Mirel), supported by grant U54 RR020278 from the National Center for Research Resources. D.R.W. and R.E.S. thank the staff of the Lieber Institute and the Clinical Brain Disorders Branch of the Intramural Research Program of the NIMH for their assistance in data collection and management. We acknowledge the Irish contribution to the International Schizophrenia Consortium (ISC) study, the WTCCC2 schizophrenia study and WTCCC2 controls from the 1958BC and UKNBS, the Science Foundation Ireland (08/IN.1/B1916). We acknowledge use of the Trinity Biobank sample from the Irish Blood Transfusion Service and the Trinity Centre for High Performance Computing. Funding for this study was provided by the WTCCC2 project (085475/B/08/Z and 085475/Z/08/Z), the Wellcome Trust (072894/Z/03/Z, 090532/Z/09/Z and 075491/Z/04/B), NIMH grants (MH 41953 and MH083094) and British 1958 Birth Cohort DNA collection funded by the MRC (grant G0000934) and the Wellcome Trust (grant 068545/Z/02). Collection of the UK National Blood Service controls was funded by the Wellcome Trust. We acknowledge Hong Kong Research Grants Council project grants GRF 774707M, 777511M, 776412M and 776513M.ENIGMA. ENIGMA was supported in part by a consortium grant (U54 EB020403 to P.M.T.) from the NIH institutes contributing to the Big Data to Knowledge (BD2K) initiative, including the NIBIB and NCI. ADNI and ADNI2GO: Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (NIH grant U01 AG024904) and US Department of Defense (award W81XWH-12-2-0012). ADNI is funded by the US National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of Southern California. Betula: this sample collection was supported by a Wallenberg Scholar grant from the Knut and Alice Wallenberg Foundation and a grant from Torsten and Ragnar Söderbergs Foundation to L.N., and a grant from HelseVest RHF (grant 911554) to S.L.H. Bipolar Family Study: The Bipolar Family Study wishes to thank the Scottish Mental Health Research Network for research assistant support; the Brain Research Imaging Centre Edinburgh, a center in the Scottish Funding Council Scottish Imaging Network–A Platform for Scientific Excellence (SINAPSE) Collaboration, for image acquisition; and the Wellcome Trust Clinical Research Facility for genotyping. Genotyping was supported by a National Alliance for Research on Schizophrenia and Depression (NARSAD) Independent Investigator Award (to A.M.M.), and data collection was supported by the Health Foundation Clinician Scientist Fellowship. BIG: This work makes use of the BIG (Brain Imaging Genetics) database, first established in Nijmegen, the Netherlands, in 2007. This resource is now part of the Cognomics Initiative (http://www.cognomics.nl/), a joint initiative by researchers of the Donders Centre for Cognitive Neuroimaging, the Human Genetics and Cognitive Neuroscience departments of the Radboud University Medical Centre and the Max Planck Institute for Psycholinguistics in Nijmegen. The Cognomics Initiative is supported by the participating departments and centers and by external grants from the Biobanking and Biomolecular Resources Research Infrastructure (Netherlands) (BBMRI-NL), the Hersenstichting Nederland and the Netherlands Organization for Scientific Research (NWO). We wish to thank all persons who kindly participated in the BIG research. The research leading to these results also receives funding from the European Community's Seventh Framework Programme (FP7/2007– 2013) under grant agreements 602450 (IMAGEMEND) and 602805 (Aggressotype) and from ERC-2010-AdG 268800-NEUROSCHEMA. B.F. is supported by a Vici grant from the NWO (grant 016.130.669). Brain Genomics Superstruct Project (GSP): Data were provided [in part] by the Brain GSP of Harvard University and the Massachusetts General Hospital, with support from the Center for Brain Science Neuroinformatics Research Group, the Athinoula A. Martinos Center for Biomedical Imaging and the Center for Human Genetic Research. Twenty individual investigators at Harvard and Massachusetts General Hospital generously contributed data to GSP. GIG: The GIG (Genomic Imaging Göttingen) sample was established at the Center for Translational Research in Systems Neuroscience and Psychiatry at Göttingen University. We thank M. Keil, E. Diekhof, T. Melcher and I. Henseler for assistance in MRI data acquisition, and E. Binder and H. Mohr for help with genotyping. We are grateful to all persons who kindly participated in the GIG study. IMAGEN: IMAGEN was supported by the European Union-funded FP6 Integrated Project IMAGEN (Reinforcement-Related Behaviour in Normal Brain Function and Psychopathology) (LSHM-CT- 2007-037286), the FP7 projects IMAGEMEND (602450) and MATRICS (603016), and the Innovative Medicine Initiative Project EU-AIMS (115300-2), the Medical Research Council programme grant “Developmental pathways into adolescent substance abuse” (93558), as well as the NIHR Biomedical Research Center “Mental Health”. Further support was provided by the Swedish Research Council (FORMAS) and the BMBF (eMED SysAlc 01ZX1311A; Forschungsnetz AERIAL; 1EV0711). MooDS: The establishment of the MooDS sample was funded by the BMBF through the Integrated Genome Research Network MooDS (grant 01GS08144 to M.M.N. and S.C., grant 01GS08147 to M.R. and A.M.-L. and grant 01GS08148 to A. Heinz), under the auspices of the National Genome Research Network plus (NGFNplus), and through the Integrated Network IntegraMent, under the auspices of the e:Med Programme (grant 01ZX1314A to M.M.N., grant 01ZX1314C to H. Walter, grant 01ZX1314G to M.R.). MPIP: The MPIP Munich Morphometry Sample comprises images acquired as part of the Munich Antidepressant Response Signature Study and the Recurrent Unipolar Depression (RUD) Case-Control study performed at the MPIP, and control subject data acquired at the Ludwig Maximilians University, Munich, Department of Psychiatry. We wish to acknowledge A. Olynyik and radiographers R. Schirmer, E. Schreiter, R. Borschke and I. Eidner for image acquisition and data preparation. We thank D.P. Auer for local study management in the initial phase of the RUD study. We are grateful to GlaxoSmithKline for providing the genotypes of the RUD case-control sample. We thank the staff of the Center of Applied Genotyping for generating the genotypes of the MARS cohort. The study is supported by a grant of the Exzellenz-Stiftung of the Max Planck Society. This work has also been funded by the BMBF in the framework of the National Genome Research Network (NGFN), FKZ 01GS0481. NCNG: Sample collection was supported by grants from the Bergen Research Foundation and the University of Bergen, the Dr Einar Martens Fund, the K.G. Jebsen Foundation and the Research Council of Norway, to S.L.H., V.M.S. and T.E. NESDA: Funding was obtained from the Netherlands Organization for Scientific Research (Geestkracht program grant 10-000-1002), the Center for Medical Systems Biology (CSMB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL), VU University's Institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam, University Medical Center Groningen, Leiden University Medical Center and NIH (R01D0042157-01A, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. Computing was supported by BiG Grid, the Dutch e-Science Grid, which is financially supported by NWO. NeuroIMAGE: NeuroIMAGE was supported by NIH grant R01MH62873 (to S.V. Faraone), NWO large investment grant 1750102007010 (to J. Buitelaar) and grants from Radboud University Nijmegen Medical Center, University Medical Center Groningen and Accare, and VU University Amsterdam. The research leading to these results also receives funding from the European Community's Seventh Framework Programme (FP7/2007– 2013) under grant agreements 602450 (IMAGEMEND), 278948 (TACTICS) and 602805 (Aggressotype). NTR-Adults and Brainscale: We would like to thank all twin participants from the Netherlands Twin Register. The NTR-Adult and Brainscale studies were supported by the NWO (MW904-61-193 (E.J.C.d.G. and D.I.B.), MaGW-nr 400-07-080 (D.v.t.E.), MagW 480-04-004 (D.B), (51.02.060 (H.H.), 668.772 (D.B. and H.H.); NWO/SPI 56-464-14192 (D.B.), the European Research Council (ERC-230374) (D.B.), High Potential Grant Utrecht University (H.H.), NWO Brain and Cognition 433-09-220 (H.H.) and the Neuroscience Campus Amsterdam. Older Australian Twins Study (OATS): We would like to acknowledge and thank the OATS participants, their supporters and respective research teams. OATS is supported by Australian National Health and Medical Research Council (NHMRC)/Australian Research Council Strategic Award 401162 and NHMRC project grant 1045325 to P.S.S. and colleagues. OATS was facilitated through access to the Australian Twin Registry, a national research resource supported by NHMRC enabling grant 310667, administered by the University of Melbourne. DNA was extracted by Genetic Repositories Australia, an Enabling Facility supported by NHMRC grant 401184. OATS genotyping was partly funded by a Commonwealth Scientific and Industrial Research Organisation Flagship Collaboration Fund grant. H.B. is supported by the Australian Government funded Dementia Collaborative Research Centre, UNSW. N.J.A. was supported by NHMRC project grant 525453 and K.A.M. is supported by an Alzheimer's Australia Dementia Research Foundation postdoctoral fellowship and NHMRC capacity building grant 568940. QTIM: D.P.H., N.J., C.R.K.C. and P.M.T. are supported, in part, by NIH grants R01 NS080655, R01AG040060, R01 EB008432, R01 MH097268, U01 AG024904, R01 MH085667, R01 MH089722, P41 EB015922 and R01 MH094343. R.K.W. is supported by National Science Foundation (BCS-1229450). J.L.S. was supported by the NIMH (K99MH102357) and Autism Speaks. S.E.M. and G.I.d.Z. are supported by Future Fellowships (FT110100548, FT0991634) from the Australian Research Council, and G.W.M. is supported by an NHMRC fellowship (619667). The QTIM study is supported by grants from the NIH (R01 HD050735) and the NHMRC (389875, 486682, 1009064). We thank the twins and siblings for their participation, M. Grace and A. Eldridge for twin recruitment, A. Al Najjar and other radiographers for scanning, K. McAloney and D. Park for research support, and A. Henders and staff for DNA sample processing and preparation. SHIP: The Study of Health in Pomerania (SHIP) is supported by the BMBF (grants 01ZZ9603, 01ZZ0103 and 01ZZ0403) and the DFG (GR 1912/5-1). Genome-wide data and MRI scans were supported by a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. SHIP-TREND-0: This cohort is part of the Community Medicine Research (CMR) net of the University of Greifswald, which is funded by the BMBF and the German Ministry of Cultural Affairs, as well as by the Social Ministry of the Federal State of Mecklenburg–West Pomerania. CMR encompasses several research projects that share data from SHIP. MRI scans were supported by a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. The SHIP authors are grateful to M. Stanke for the opportunity to use his server cluster for SNP imputation as well as to H. Prokisch and T. Meitinger (HelmholtzZentrum München) for genotyping the SHIP-TREND cohort, which was supported by the BMBF (grant 03ZIK012). We thank all staff members and participants of the SHIP studies, as well as all of the genotyping staff for generating the SHIP SNP data set. D.J. is supported by a scholarship from the Gerhard-Domagk Programme of the University Medicine Greifswald. Sydney Memory and Ageing Study (Sydney MAS): We would like to thank the Sydney MAS participants, their supporters and respective research teams. Sydney MAS was supported by NHMRC program grants 350833 and 568969 to P.S.S., H.B. and G. Andrews. DNA was extracted by Genetic Repositories Australia, an Enabling Facility supported by NHMRC grant 401184. H.B. is supported by the Australian Government funded Dementia Collaborative Research Centre, UNSW. N.J.A. was supported by NHMRC project grant 525453 and K.A.M. is supported by an Alzheimer's Australia Dementia Research Foundation postdoctoral fellowship. Both S. Reppermund and K.A.M. are supported by NHMRC capacity building grant 568940. Data used in preparing this article were obtained from the ADNI database (http://adni.loni.usc.edu/). Many investigators in ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Author information
Authors and Affiliations
Consortia
Contributions
Study conception and supervision: B. Franke, B.M.N., J.L.S., M.C.O'D., P.F.S., P.M.T., S.E.M. Design of ENIGMA or PGC: A.A.-V., A.M.M., B. Franke, B.M.N., D.P.H., J.A.T., J.L.S., J.W.S., K.J.E.v.H., M.C.N., M.C.O'D., O.A.A., P.F.S., P.L., P.M.T., S.E.M., S. Ripke, T.E.N., V.A., Y.Y., Y.Y.W.H. Obtained funding: B. Franke, M.C.O'D., M.J.W., N.G.M., P.F.S., P.M.T. Provided samples: PGC2 Schizophrenia Working Group and ENIGMA2 Consortium. Conducted analyses: A.A.-V., B.M.N., D.P.H., J.L.S., K.J.E.v.H., M.C.N., P.L., S.E.M., S. Ripke, T.E.N., V.A., Y.Y.W.H. Writing group: A.A.-V., B. Franke, B.M.N., D.P.H., J.L.S., K.J.E.v.H., M.C.O'D., P.F.S., P.M.T., S.E.M., S. Ripke, V.A. All authors reviewed and approved the final version of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
Some of the authors or collaborators were employees of the following pharmaceutical companies: Pfizer (C.R.S., J.R.W., H.S.X.), F. Hoffman-La Roche (E. Domenici, L.E.), Eli Lilly (D.A.C., Y. Mokrab, L. Nisenbaum) and Janssen (S. Gopal, D. Wang, Q.S.L., N. Cohen). None of these companies influenced the design of the study, the interpretation of the data or the amount of data reported, or financially profit by publication of the results, which are precompetitive.
Additional information
Psychosis Endophenotypes International Consortium, London, UK
Integrated supplementary information
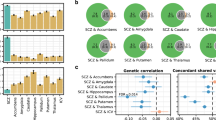
Supplementary Figure 1 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA nucleus accumbens volume
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENIGMA MA is shown in the lower panel.
Supplementary Figure 2 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA amygdala volume.
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENIGMA MA is shown in the lower panel.
Supplementary Figure 3 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA caudate nucleus volume.
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENIGMA MA is shown in the lower panel.
Supplementary Figure 4 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA hippocampus volume.
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENIGMA MA is shown in the lower panel.
Supplementary Figure 5 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA ICV.
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENGIMA MA is shown in the lower panel.
Supplementary Figure 6 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA pallidum volume.
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENIGMA MA is shown in the lower panel.
Supplementary Figure 7 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA putamen volume.
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENGIMA MA is shown in the lower panel.
Supplementary Figure 8 P-value-based meta-analysis between PGC schizophrenia (SCZ) and ENIGMA thalamus volume.
PGC SCZ MA is shown in the top panel, the MA between PGC and ENIGMA is shown in the middle panel, and the ENIGMA MA is shown in the lower panel.
Supplementary Figure 9 Conjunction analysis: regional plots of genetic loci that influence both risk for schizophrenia and brain structure
We used a conjunction test to identify specific genetic variants influencing both risk for schizophrenia and changes in brain structure genome-wide. The conjunction test was run genome-wide using 7,510,842 SNPs found in both the ENIGMA2 and PGC2 studies with MAF ≥ 0.01. Conjunction results are then corrected for a downward bias in significance using the relaxed intersection union test. No SNP demonstrated genome-wide significant association to both schizophrenia and brain structure. Several sub-threshold loci did arise from this analysis that bear comment given the possibility that stronger significance may be achieved in future studies. (a) First, the conjunction between intracranial volume and schizophrenia was associated with a locus on chromosome 6 marked by rs381349, where the T allele increases ICV and decreases schizophrenia risk, in the expected direction for a schizophrenia risk factor. This locus is found intronic in the FOXO3 gene known to be involved in neural stem cell proliferation and renewal. (b) The conjunction between hippocampal volume and schizophrenia was associated with a previously mentioned locus on chromosome 2 marked by rs11693702, although in the opposite direction expected for a schizophrenia risk factor (A allele increases schizophrenia risk and increases hippocampal volume). (c,d) The conjunction between putamen volume and schizophrenia was associated with two interesting loci in the expected direction for schizophrenia risk factors marked by rs958246 (chromosome 11; A allele is associated with increased schizophrenia risk and increased putamen volume) and rs7231178 (chromosome 18; A allele is associated with increased schizophrenia risk and increased putamen volume). These SNPs are found in intronic regions of the DLG2 gene, which encodes a key component of the post-synaptic density, and the DCC gene, a netrin receptor involved in axon guidance.
Supplementary Figure 10 Comparison of SNP effect sizes
Effect sizes of individual SNPs having achieved replicated evidence for association with the phenotypes indicated. For brain structure phenotypes and schizophrenia, effect sizes for both, discovery and replication analyses are shown. For schizophrenia, only the five top SNPs from the 128 recently described genome-wide significant findings were included in the comparison. Height and educational attainment are shown as additional comparators. Effect sizes were measured in percent variance explained (for quantitative traits) or percent variance explained on the liability scale (for disease categories).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Table 4 (PDF 2300 kb)
Supplementary Table 1: Characteristics of the PGC schizophrenia and ENIGMA data sets
Tag refers to the PGC sample identifier. The numbers shown are before exclusion of overlap with ENIGMA. (XLSX 12 kb)
Supplementary Table 2: Reciprocal look-up of genome-wide significant findings in the other data set
Panels 1–8: Shown are LD-independent genome-wide significant SNP associations for schizophrenia from the final analysis (sorted by genomic position according to UCSC hg19/NCBI Build 37). Markers are ranked from 1 to 128 in order of significance. Insertion/deletion variants are given in the form “chrA_B_C” where A = chromosome, B = position, and C = insertion (I) or deletion (D). Column A12 has the SNP alleles, with the first allele (a1) the reference allele for the frequency and odds ratio columns. Frq = frequency of allele 1. Chr and Position denote the associated region surrounding the index SNP containing 1 or more SNPs in LD (r2 > 0.6) with the index SNP. OR = odds ratio for allele 1, CI = 95% confidence interval for OR. Effect = effect size for allele 1, CI = 95% confidence interval for the effect size and are provided in units of mm3 per effect allele. †Top finding from schizophrenia (rs115329265) was not available in ENIGMA and was replaced by a SNP in moderate LD (6:28305863R; r2 = 0.64). *X-chromosome-located SNPs were excluded because X-chromosomal information was not available in ENIGMA. **rs77149735 was not available in ENIGMA and could not be replaced by a SNP in moderate or high LD. Panel 9: The allele frequency (Freq) and effect size (Effect) are given with reference to allele 1 (A1). CI = 95% confidence interval for the effect size and is provided in units of mm3 per effect allele. OR = odds ratio for allele 1, CI = 95% confidence interval for OR. (XLSX 116 kb)
Supplementary Table 3: P-values for the PGC SCZ and ENIGMA volumes and combined meta-analyses
The combined meta-analysis results for each brain volume was ‘clumped’ with the 1000 Genomes V3 reference panel for Europeans using PLINK. P-value-informed LD clumping was done using 5 × 10-8 as significance threshold for index SNPs, 5 × 10-6 as secondary significance threshold for clumped SNPs, 0.8 as LD r2 threshold, and 1 MB as physical distance threshold. (XLSX 245 kb)
Rights and permissions
About this article
Cite this article
Franke, B., Stein, J., Ripke, S. et al. Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat Neurosci 19, 420–431 (2016). https://doi.org/10.1038/nn.4228
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4228
This article is cited by
-
Effect of schizophrenia common variants on infant brain volumes: cross-sectional study in 207 term neonates in developing Human Connectome Project
Translational Psychiatry (2023)
-
Identifying genes associated with brain volumetric differences through tissue specific transcriptomic inference from GWAS summary data
BMC Bioinformatics (2022)
-
Genome-Wide association study of quantitative biomarkers identifies a novel locus for alzheimer’s disease at 12p12.1
BMC Genomics (2022)
-
Mapping the genetic architecture of cortical morphology through neuroimaging: progress and perspectives
Translational Psychiatry (2022)
-
Derivation and utility of schizophrenia polygenic risk associated multimodal MRI frontotemporal network
Nature Communications (2022)