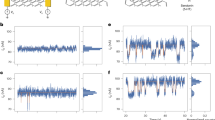
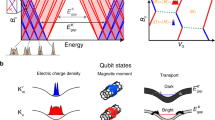
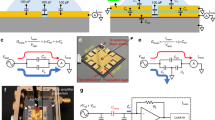
Abstract
Single-molecule measurements of biomolecules can provide information about the molecular interactions and kinetics that are hidden in ensemble measurements. However, there is a requirement for techniques with improved sensitivity and time resolution for use in exploring biomolecular systems with fast dynamics. Here, we report the detection of DNA hybridization at the single-molecule level using a carbon nanotube field-effect transistor. By covalently attaching a single-stranded probe DNA sequence to a point defect in a carbon nanotube, we are able to measure two-level fluctuations in the conductance of the nanotube in the presence of a complementary DNA target. The kinetics of the system are studied as a function of temperature, allowing the measurement of rate constants, melting curves and activation energies for different sequences and target concentrations. The kinetics demonstrate non-Arrhenius behaviour, in agreement with DNA hybridization experiments using fluorescence correlation spectroscopy. This technique is label-free and could be used to probe single-molecule dynamics at microsecond timescales.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bonnet, G., Krichevsky, O. & Libchaber, A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA 95, 8602–8606 (1998).
Li, H. T., Ren, X. J., Ying, L. M., Balasubramanian, S. & Klenerman, D. Measuring single-molecule nucleic acid dynamics in solution by two-color filtered ratiometric fluorescence correlation spectroscopy. Proc. Natl Acad. Sci. USA 101, 14425–14430 (2004).
Wallace, M. I., Ying, L., Balasubramanian, S. & Klenerman, D. Non-Arrhenius kinetics for the loop closure of a DNA hairpin. Proc. Natl Acad. Sci. USA 98, 5584–5589 (2001).
Deniz, A. A., Mukhopadhyay, S. & Lemke, E. A. Single-molecule biophysics: at the interface of biology, physics and chemistry. J. R. Soc. Interface 5, 15–45 (2008).
Fei, J., Kosuri, P., MacDougall, D. D. & Gonzalez, R. L. Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 30, 348–359 (2008).
Patolsky, F. et al. Electrical detection of single viruses. Proc. Natl Acad. Sci. USA 101, 14017–14022 (2004).
Armani, A. M., Kulkarni, R. P., Fraser, S. E., Flagan, R. C. & Vahala, K. J. Label-free, single-molecule detection with optical microcavities. Science 317, 783–787 (2007).
Burg, T. P. et al. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 446, 1066–1069 (2007).
Hughes, R. C., Ricco, A. J., Butler, M. A. & Martin, S. J. Chemical microsensors. Science 254, 74–80 (1991).
Cecconi, C., Shank, E. A., Bustamante, C. & Marqusee, S. Direct observation of the three-state folding of a single protein molecule. Science 309, 2057–2060 (2005).
Besteman, K., Lee, J. O., Wiertz, F. G. M., Heering, H. A. & Dekker, C. Enzyme-coated carbon nanotubes as single-molecule biosensors. Nano Lett. 3, 727–730 (2003).
Star, A. et al. Label-free detection of DNA hybridization using carbon nanotube network field-effect transistors. Proc. Natl Acad. Sci. USA 103, 921–926 (2006).
Guo, X., Gorodetsky, A. A., Hone, J., Barton, J. K. & Nuckolls, C. Conductivity of a single DNA duplex bridging a carbon nanotube gap. Nature Nanotech. 3, 163–167 (2008).
Goldsmith, B. R. et al. Conductance-controlled point functionalization of single-walled carbon nanotubes. Science 315, 77–81 (2007).
Goldsmith, B. R., Coroneus, J. G., Kane, A. A., Weiss, G. A. & Collins, P. G. Monitoring single-molecule reactivity on a carbon nanotube. Nano Lett. 8, 189–194 (2008).
Ishida, M., Hongo, H., Nihey, F. & Ochiai, Y. Diameter-controlled carbon nanotubes grown from lithographically defined nanoparticles. Jpn J. Appl. Phys. 2 43, L1356–L1358 (2004).
Chen, R. J. et al. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proc. Natl Acad. Sci. USA 100, 4984–4989 (2003).
Kosynkin, D. V. et al. Longitudinal unzipping of carbon nanotubes to form graphene nanoribbons. Nature 458, 872–876 (2009).
Bachtold, A. et al. Scanned probe microscopy of electronic transport in carbon nanotubes. Phys. Rev. Lett. 84, 6082–6085 (2000).
Freitag, M. et al. Imaging of the Schottky barriers and charge depletion in carbon nanotube transistors. Nano Lett. 7, 2037–2042 (2007).
Minot, E. D. et al. Carbon nanotube biosensors: the critical role of the reference electrode. Appl. Phys. Lett. 91, 093507 (2007).
Borer, P. N., Dengler, B., Tinoco, I. Jr & Uhlenbeck, O. C. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 86, 843–853 (1974).
Gong, P. & Levicky, R. DNA surface hybridization regimes. Proc. Natl Acad. Sci. USA 105, 5301–5306 (2008).
Sun, Y., Harris, N. C. & Kiang, C. H. Melting transition of directly linked gold nanoparticle DNA assembly. Physica A 350, 89–94 (2005).
Karachevtsev, V. A. et al. Adsorption of poly(rA) on the carbon nanotube surface and its hybridization with poly(rU). ChemPhysChem 9, 2010–2018 (2008).
Brewood, G. P. et al. Electrical detection of the temperature induced melting transition of a DNA hairpin covalently attached to gold interdigitated microelectrodes. Nucleic Acids Res. 36, e98 (2008).
Bronson, J. E., Fei, J., Hofman, J. M., Gonzalez, R. L. Jr & Wiggins, C. H. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys J. 97, 3196–3205 (2009).
Fei, J. et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc. Natl Acad. Sci. USA 106, 15702–15707 (2009).
Chan, V., Graves, D. J. & McKenzie, S. E. The biophysics of DNA hybridization with immobilized oligonucleotide probes. Biophys J. 69, 2243–2255 (1995).
von Hippel, P. H. & Berg, O. G. Facilitated target location in biological systems. J. Biol. Chem. 264, 675–678 (1989).
Halford, S. E. & Marko, J. F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32, 3040–3052 (2004).
Ansari, A., Kuznetsov, S. V. & Shen, Y. Configurational diffusion down a folding funnel describes the dynamics of DNA hairpins. Proc. Natl Acad. Sci. USA 98, 7771–7776 (2001).
Oliveberg, M., Tan, Y. J. & Fersht, A. R. Negative activation enthalpies in the kinetics of protein folding. Proc. Natl Acad. Sci. USA 92, 8926–8929 (1995).
Dobson, C. M., Sali, A. & Karplus, M. Protein folding: a perspective from theory and experiment. Angew. Chem. Int. Ed. 37, 868–893 (1998).
Chalikian, T. V., Volker, J., Plum, G. E. & Breslauer, K. J. A more unified picture for the thermodynamics of nucleic acid duplex melting: a characterization by calorimetric and volumetric techniques. Proc. Natl Acad. Sci. USA 96, 7853–7858 (1999).
Altan-Bonnet, G., Libchaber, A. & Krichevsky, O. Bubble dynamics in double-stranded DNA. Phys. Rev. Lett. 90, 138101 (2003).
Metzler, R., Ambjornsson, T., Hanke, A. & Fogedby, H. C. Single DNA denaturation and bubble dynamics. J. Phys. Condens. Matter 21, 034111 (2009).
Huang, L. M., Cui, X. D., White, B. & O'Brien, S. P. Long and oriented single-walled carbon nanotubes grown by ethanol chemical vapor deposition. J. Phys. Chem. B 108, 16451–16456 (2004).
Ishigami, M., Chen, J. H., Cullen, W. G., Fuhrer, M. S. & Williams, E. D. Atomic structure of graphene on SiO2 . Nano Lett. 7, 1643–1648 (2007).
Hermanson, G. T. Bioconjugate Techniques 2nd edn (Academic Press, 2008).
Acknowledgements
This work was supported in part by the National Science Foundation (grants ENG-0707748 and CHE-0641523). Additional support was provided by the New York State Office of Science, Technology, and Academic Research (NYSTAR). This work was also supported in part by the Office of Naval Research (grants N00014-09-01-0250 and N00014-09-1-1117) and by the National Institutes of Health (grant R33-HG003089).
Author information
Authors and Affiliations
Contributions
S.S., C.-Y.C. and K.L.S. designed the experiments. S.S. and C.-Y.C. performed the experiments and analysed the data. Y.-J.Y. and P.K. assisted in the AFM and SGM experiments. C.N and R.L.G. assisted with data analysis. S.S., C.-Y.C and K.L.S. co-wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 2758 kb)
Rights and permissions
About this article
Cite this article
Sorgenfrei, S., Chiu, Cy., Gonzalez, R. et al. Label-free single-molecule detection of DNA-hybridization kinetics with a carbon nanotube field-effect transistor. Nature Nanotech 6, 126–132 (2011). https://doi.org/10.1038/nnano.2010.275
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2010.275