Abstract
Heavy cannabis users display smaller amygdalae and hippocampi than controls, and genetic variation accounts for a large proportion of variance in liability to cannabis dependence (CD). A single nucleotide polymorphism in the cannabis receptor-1 gene (CNR1), rs2023239, has been associated with CD diagnosis and intermediate phenotypes, including abstinence-induced withdrawal, cue-elicited craving, and parahippocampal activation to cannabis cues. This study compared hippocampal and amygdalar volumes (potential CD intermediate phenotypes) between heavy cannabis users and healthy controls, and analyzed interactions between group, rs2023239 variation, and the volumes of these structures. Ninety-four heavy cannabis users participated, of whom 37 (14 men, 23 women; mean age=27.8) were matched to 37 healthy controls (14 men, 23 women; mean age=27.3) for case-control analyses. Controlling for total intracranial volume and other confounding variables, matched cannabis users had smaller bilateral hippocampi (left, p=0.002; right, p=0.001) and left amygdalae (p=0.01) than controls. When genotype was considered in the case-control analyses, there was a group by genotype interaction, such that the rs2023239 G allele predicted lower volume of bilateral hippocampi among cannabis users relative to controls (both p<0.001). This interaction persisted when all 94 cannabis users were compared to controls. There were no group by genotype interactions on amygdalar volume. These data replicate previous findings of reduced hippocampal and amygdalar volume among heavy cannabis users, and suggest that CNR1 rs2023239 variation may predispose smaller hippocampal volume after heavy cannabis use. This association should be tested in future studies of brain volume differences in CD.
Similar content being viewed by others
INTRODUCTION
Cannabis is the most frequently used illicit substance, and questions have arisen regarding its effects on brain structure and function (Iversen, 2003). Structural neuroimaging studies have indicated that volumes of several brain areas are smaller in heavy cannabis users (CU) (Ashtari et al, 2011; Demirakca et al, 2011; Matochik et al, 2005; Wilson et al, 2000; Yucel et al, 2008). Across studies, reductions in hippocampal and amygdalar volumes are most consistent (Lorenzetti et al, 2010). Of these structures, hippocampal volume appears to be particularly sensitive to heavy cannabis use. Among heavy CU, hippocampal volume has been negatively correlated with duration and amount of lifetime (Ashtari et al, 2011; Yucel et al, 2008) and recent (Cousijn et al, 2012) use. Further, right hippocampal gray matter volume and bilateral hippocampal gray matter concentration have been inversely associated with the ratio of delta-9-tetrahydrocannabinol (Δ9-THC), the primary psychoactive component in cannabis, to cannabidiol, a cannabinoid compound whose psychoactive effects may oppose those of Δ9-THC (Bhattacharyya et al, 2010), in hair samples taken from heavy CU, suggesting that hippocampal effects are more likely among individuals who use cannabis with greater relative Δ9-THC content (Demirakca et al, 2011).
Genetic variation is strongly related to the initiation and maintenance of heavy cannabis use. Heritability estimates range from 17–72% for having ever used cannabis, and from 33–76% for cannabis dependence (CD) (Agrawal and Lynskey, 2006), which is characterized by the development of cannabis tolerance and withdrawal, spending a great deal of time obtaining and using cannabis, using it in larger amounts or for longer periods than intended, continuing use despite negative consequences, and unsuccessful efforts to control use. One candidate gene for CD, CNR1, encodes for the type 1 cannabinoid receptor (CB1), a presynaptically expressed Gi/Go-protein-coupled receptor that is densely localized to the hippocampus, amygdala, prefrontal cortex (PFC), striatum, and cerebellum (Herkenham et al, 1991). Δ9-THC binds to CB1 receptors and, acting as a retrograde signaling molecule, enacts a variety of second-messenger signaling cascades (Pazos et al, 2005; Szabo et al, 1998). Of particular relevance to the development of addictive behavior, CB1 receptors on GABAergic interneurons in the ventral tegmental area disinhibit the mesocorticolimbic dopaminergic pathways and increase dopamine release in the nucleus accumbens (Cheer et al, 2004; Gessa et al, 1998; Szabo et al, 2002), a mechanism common to most drugs of abuse. CNR1 variation has been associated with CD diagnosis in several large samples (Agrawal et al, 2009; Hartman et al, 2009; Hopfer et al, 2006), as well as with other diagnostic substance use disorder (SUD) phenotypes (for review see Benyamina et al, 2011).
Variants that have demonstrated association with CD and SUD include seven single nucleotide polymorphisms (SNPs) (rs806368, rs806379, rs806380, rs1049353, rs2023239, rs6454674, and rs12720071) and a triplet repeat polymorphism ((AAT)n). A recent meta-analysis suggested the AATn repeat had the most consistent association with substance dependence diagnosis (Benyamina et al, 2011). However, association studies have not been well replicated, perhaps in part due to their reliance on dichotomous diagnostic phenotypes and the unknown functional relevance of some of the variants (Agrawal and Lynskey, 2009). In contrast, our own prior work has focused on rs2023239, a SNP that may affect CB1 expression. This SNP encodes an A to G substitution in the third exon of CNR1 that appears to engender an alternative splice variant of the gene (Zhang et al, 2004). We have studied its association with CD intermediate phenotypes, which may more closely reflect the neurobiology of this disorder than diagnostic phenotypes. In human postmortem brain tissue, the rs2023239 G allele is associated with greater CB1 expression in PFC (Hutchison et al, 2008). Among heavy CU, this allele is also associated with greater withdrawal and craving for cannabis after short-term abstinence, greater craving after cannabis cue exposure (Haughey et al, 2008), and greater brain response to cannabis cues in areas including the hippocampus, anterior cingulate, and medial PFC (Filbey et al, 2010).
Given these findings, we investigated the effects of rs2023239 variance on hippocampal and amygdalar volumes, which may represent an intermediate phenotype for SUD (Benegal et al, 2007), among heavy CU and healthy controls (HC). The current study had two aims: (1) to compare the volumes of these structures between heavy CU and HC; and (2) to analyze interactions between group, rs2023239 variation, and the volumes of these structures.
MATERIALS AND METHODS
Participants
Ninety-four regular CU and 37 HC who had never used cannabis regularly were recruited from the greater Albuquerque, NM metro area. Thirty-seven CU were matched to HC on the basis of gender, age, and years of education. After matching gender, the CU with the closest correspondence in (1) age and (2) education was selected. Rs2023239 genotype did not influence selection. We previously reported on cannabis cue-elicited brain activation among a subset of the CU (Filbey et al, 2009, 2010). The 37 matched CU-HC pairs were used for the primary analyses. Because we anticipated that only 30% of individuals would carry at least one copy of the rs2023239 minor (G) allele, we also sought to determine whether genotype effects were present when the entire sample of CU, which included more G-allele carriers, was considered. Hereafter, the 94 CU are referred to as ‘all CU,’ and the 37 CU matched to HC as ‘matched CU.’
Inclusion/Exclusion Criteria
All participants were required to be ages 18–50, right-handed, English-speaking, free of neurological disorders and psychotic symptoms, including all SUD (except nicotine dependence and, for CU, cannabis use disorders), as diagnosed by the Psychotic Symptoms and Substance Use Disorders modules of the Structured Clinical Interview for DSM-IV-TR, Research Version (First et al, 2002), and free of magnetic resonance imaging (MRI) contraindications (ie, metallic implants, claustrophobia, pregnancy). CU were required to report using cannabis at least 4 times per week over the past 6 months, while HC were required to report never having used cannabis regularly or other illicit substances in the past 6 months. Some CU reported recent infrequent (monthly or less) use of other illicit substances: specifically, 24 CU reported hallucinogen use (of whom six were matched CU), three reported ecstasy use, two reported cocaine use, and one reported methamphetamine use (none of those who reported ecstasy, cocaine, or methamphetamine use were matched CU).
Procedure
The University of New Mexico Human Research Review Committee approved all procedures. The study comprised two sessions separated by three days, during which participants were instructed to abstain from cannabis use (this provision was intended to increase reactivity to cannabis cues during the second session). At the first session, after providing informed consent, participants were assessed and provided saliva and urine samples for DNA and urine drug analyses, respectively. At the second session, participants provided another urine sample and underwent the MRI scan. All CUs’ urine was positive for Δ9-THC (cut-off: 50 ng/ml) and negative for other substances at both sessions; all HCs’ urine was negative for all substances. Participants were compensated for their participation.
Assessment
Demographic information was assessed with a questionnaire. Race/ethnicity category options were: Caucasian; African-American; Asian-American; Latino; and Native American. Past 90-day cannabis, alcohol, and nicotine were assessed with the Timeline Follow-back (TLFB), a researcher-administered, calendar-assisted interview (Sobell and Sobell, 1992). IQ was assessed with the Wechsler Abbreviated Scale of Intelligence (WASI; Wechsler, 1999). Trained research assistants administered both measures. The TLFB was not administered to 12 HC; these individuals reported drinking fewer than 14 drinks per week (seven for women) during a phone screen, but more detailed drinking data were not available. Lifetime cannabis use was assessed with a questionnaire, and depressive symptoms were assessed with the Beck Depression Inventory, 2nd Edition (BDI-II; Beck et al, 1996). Participants were considered cigarette smokers if they reported smoking ⩾10 cigarettes/day in the past 90 days. Demographic information, WASI and BDI-II scores, substance use history, total intracranial volume (ICV), and raw hippocampal and amygdalar volumes for HC and matched CU are listed in Table 1 by rs2023239 genotype. The same information for all CU is listed in Table 2.
Brain Imaging
Imaging was conducted with a 3T TIM TRIO scanner (Siemens, Erlangen, Germany) and a multi-echo magnetization prepared rapid gradient echo (MPRAGE) sequence with the following parameters: repetition/echo/inversion recovery times=2300/2.74/900 ms, flip angle=81°, field of view=256 × 256 mm, slab thickness=176 mm, matrix=256 × 256 × 176, voxel size=1 × 1 × 1 mm, number of echoes=4, pixel bandwidth=650 Hz, total scan time=6 min. Before volumetric analysis, images were inspected to ensure they were free of motion or obvious pathology.
Volumetric Analysis
Hippocampal and amygdalar volumes were computed with the FreeSurfer image analysis suite, v4.50 (http://surfer.nmr.mgh.harvard.edu) (Fischl et al, 2004), which performs automated cortical reconstruction and subcortical segmentation by using a probabilistic atlas to assign one of 40 different tissue class labels to every voxel in the brain. Briefly, this processing includes non-brain tissue removal, automated Talairach transformation, subcortical gray/white matter segmentation, intensity normalization, tessellation of the gray matter/white matter boundary, automated topology correction, and surface deformation following intensity gradients between gray/white and gray/cerebrospinal fluid borders. Total ICV was calculated using the atlas normalization procedure implemented in FreeSurfer, which correlates highly (r>0.9) with manual ICV measurement (Buckner et al, 2004).
Genotyping
Genomic DNA was collected from saliva and extracted according to modified standard procedures (Walker et al, 1999). Rs2023239 genotype was determined with an Applied Biosystems (Life Technologies, Carlsbad, CA) thermocycler and a Taqman (Invitrogen, Carlsbad, CA) polymerase chain reaction assay, run in triplicate. Following convention for this SNP (eg, Herman et al, 2006), individuals with at least one copy of the G (minor) allele were compared to A/A homozygotes. The rarity of the G allele precluded an additive analysis for the effects of this allele.
Statistical Analysis
Between-groups comparisons of demographic and substance use variables and ICV were conducted with χ2 tests and independent samples Student's t-tests. When variances were not equal between groups (Levene's test), unequal variances (Welch's) t-tests were used.
To analyze the main effect of group (CU vs HC) on hippocampal and amygdalar volumes, the 37 matched pairs were compared using hierarchical linear modeling (HLM) (Raudenbush and Bryk, 2002). HLM was used for this analysis because of the paired case-control design; by treating individuals as nested pairs, HLM allows more robust estimation of between-groups differences than the general linear model (GLM) or analysis of variance. In each of four HLMs, at the first level, the volume of the right or left hippocampus or amygdala was entered as the dependent variable, and group (indicating whether the subject in each pair was the CU or HC) was entered as the independent variable. Because hippocampal and amygdalar volumes scale with total ICV, this variable was entered as a first-level, group-centered covariate, as was number of cigarettes per day (to control for tobacco use). At the second level, the intercept of each first-level model was allowed to vary randomly, permitting each pair to have a different mean volume. Full maximum likelihood estimation with robust standard errors was used. To control for depressive symptoms and alcohol use, these models were also run with BDI-II score included as a first-level covariate, as well as with drinks per drinking day and drinking days per month included for the 25 pairs of subjects for whom full drinking data were available.
To analyze the effects of group, genotype, and their interaction on hippocampal and amygdalar volumes, the GLM was used to compare both the matched pairs (among whom rs2023239 genotype was not identically distributed, precluding use of HLM) and all CU vs the 37 HC. Total ICV was covaried in all models, and variables on which the groups differed (ie, age, gender, education, and number of cigarettes per day) were covaried. To control for depressive symptoms and alcohol use, these models were also run with BDI-II scores, drinks/drinking day, and drinking days/month included as covariates.
To analyze associations among all CU between hippocampal and amygdalar volumes and years of regular cannabis use, age of first cannabis use, and IQ, partial correlation coefficients were computed, controlling for gender, education, total ICV, BDI-II score, cigarettes/day, drinks/drinking day, and drinking days/month. For all analyses, an α level of p<0.05 was used. Effect sizes are listed as Cohen's d for main and simple effects and partial η2 for interactions.
RESULTS
Sample Characteristics
CU reported using cannabis, on average, more than six days per week. Most CU (33/37 of matched CU and 85/94 of all CU) reported using cannabis daily, with an average of nearly four uses per day (matched CU, M=3.9 (SD=3.5); all CU, M=3.7 (2.7)). Matched CU reported having used cannabis regularly for slightly longer than all CU, but this difference was not significant when age was covaried, suggesting it derived from the fact that matched CU were slightly older than all CU. Both matched and all CU reported significantly more alcohol use (drinks/drinking day and drinking days/month) than HC, and were more likely to be cigarette smokers. Additionally, all CU were, on average, significantly younger, more male, and less educated than HC, and had larger ICV, although there were no between-groups differences in ICV when gender was covaried (F(1, 128)=0.001, ns). Some data were not available for all participants; specifically, WASI scores were missing for one HC and two CU (of whom one was a matched CU), and BDI-II scores were missing for three CU (of whom two were matched CU).
Integrity of Volumetric and Genetic Data
Hippocampal and amygdalar volumes were normally distributed in all groups, and other assumptions of linear modeling (ie, normality, independence, and homoscedasticity of errors) were met. For rs2023239, the HapMap allele frequency for the G allele for a group of European descent is 0.167, yielding expected genotype frequencies of 0.028 for G/G, 0.278 for A/G, and 0.693 for A/A. Consistent with these expected frequencies, across all participants, actual genotype frequencies were 0.008 for G/G (one participant, a CU), 0.344 for A/G, and 0.649 for A/A, and did not differ by racial or ethnic group. Among both HC and CU, the SNP was in Hardy-Weinberg equilibrium. Allele frequency was not different between HC and matched CU (χ2(1, N=74)=0.53, p=0.47), nor between HC and all CU (χ2(1, N=131)=0.084, p=0.77).
Matched CU vs HC
HLM results indicated that matched CU had smaller bilateral hippocampi (left, t(70)=−3.35, p=0.002, d=0.71; right, t(70)=−3.45, p=0.001, d=0.73) and left amygdalae (t(70)=−2.55, p=0.01, d=0.50) than HC. On average, left and right hippocampi were 6.9 and 7.1% smaller, respectively, and left amygdala was 5.8% smaller among CU than among HC. The difference in right amygdalar volume between groups (5.6% smaller among CU) was in the same direction but not significant (t(70)=−1.86, p=0.07, d=0.41) (see Figure 1). These effects remained at the same level of significance after BDI-II scores and drinking variables were covaried.
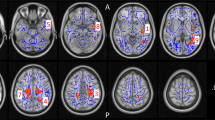
Bilateral hippocampal and amygdalar volumes for cannabis users (CU) and matched healthy controls (HC). Data are means from the HLM for each structure (±1 standard error of the mean), controlling for ICV and tobacco use. The axial slice at top right shows the FreeSurfer-delineated amygdala (light blue) and hippocampus (yellow) from a single participant (the reader is referred to the online version for references to color). **p⩽0.01 for difference between CU and HC.
Genotype Effects
For bilateral hippocampi, GLM results indicated a significant group by genotype interaction when matched CU were compared to HC, and also when all CU were compared to HC (Figure 2 displays volumes for matched CU vs HC, and Table 3 displays GLM statistics for interactions and simple effects). For the matched samples, the simple effect of group was significant in G-allele carriers, such that CU had smaller bilateral hippocampal volumes (left, 14.6% smaller; right, 13.4% smaller) than HC. Among A-allele homozygotes, there were no significant differences. The simple effect of genotype was significant among HC for left hippocampus, such that G-allele carriers had 6.8% larger volumes than A-allele homozygotes, and approached significance in the opposite direction among CU, such that G-allele carriers had 6.3% smaller volumes. For right hippocampus, simple genotype effects were in the same direction but not significant among CU or HC. Significant simple effects were similar for the analysis of all CU vs HC. These effects also remained at the same level of significance after BDI-II score and drinking variables were covaried.
Bilateral hippocampal volumes for cannabis users and matched healthy controls, by CNR1 rs2023239 genotype. Data are estimated marginal means from the GLM for each structure (±1 standard error of the mean), controlling for ICV, gender, age, education, and tobacco use. *p⩽0.05 for interaction between group and genotype; **p⩽0.05 for difference between HC A-allele homozygotes and G-allele carriers; ***p⩽0.001 for difference between CU and HC G-allele carriers.
For left amygdala, there was a significant main effect of group, such that CU had smaller volumes, but no significant group by genotype interaction or main effect of genotype. For right amygdala, there were neither significant main effects of group or genotype nor a group by genotype interaction.
Associations with Other Variables
Among all CU, controlling for the confounding variables listed above, partial correlations between hippocampal and amygdalar volumes and years of regular cannabis use ranged from −0.03 to 0.10, with all p values>0.35. Partial correlations between these volumes and age of first regular use ranged from −0.19 to −0.10, with all p values>0.05. None of these correlations differed by rs2023239 genotype. Age was not covaried in these analyses because it was strongly correlated with both years of regular use (r(94)=0.93, p<0.001) and, interestingly, age of first regular use (r(94)=0.29, p=0.005), suggesting that younger CU reported having started using cannabis regularly at an earlier age. To determine whether years of regular use or age of first regular use mediated the genotype by group interactions reported above, we tested mediation models among all CU in which each variable was added as a predictor to a GLM that regressed hippocampal volume on genotype, and found no evidence for mediation (ie, neither variable significantly predicted hippocampal volume or reduced the magnitude of the beta weight for genotype). Since there were also no differences between A- and G-allele-carrying CU on years of regular use (independent samples t-test, t(92)=−0.343, p=0.73) or age of first regular use (t(92) −0.475, p=0.64), there was no evidence that the G allele differentially predisposed early initiation or long-term maintenance of heavy cannabis use. Hippocampal and amygdalar volumes also did not have significant partial correlations with WASI full-scale IQ (p>0.20).
DISCUSSION
This study tested hippocampal and amygdalar volume differences between heavy CU and matched HC, as well as interactions between these groups and variance at the rs2023239 CNR1 SNP on the volumes of these structures. Overall, there were medium-sized differences between CU and HC in hippocampal and amygdalar volumes, and the presence of the rs2023239 G allele moderated the hippocampal differences, with a large difference between HC and CU among G-allele carriers. These findings suggest that a common genetic variant may moderate volume differences in medial temporal structures related to heavy cannabis use.
These data replicate previous findings of reduced hippocampal and amygdalar volume among heavy CU, relative to HC (Ashtari et al, 2011; Demirakca et al, 2011; Matochik et al, 2005; Wilson et al, 2000; Yucel et al, 2008). These structures may be particularly sensitive to cannabis effects because of their density of CB1 expression (Herkenham et al, 1991; Yucel et al, 2008). The hippocampus, a structure critical to learning and memory that is tightly interconnected with reward-related areas including the striatum, orbitofrontal cortex, and insula, underlies many aspects of addictive behavior, including encoding and retrieval of drug-related memories (Robbins et al, 2008), conditioned place preference (Takano et al, 2010), and context-dependent drug sensitization (Xia et al, 2011). In animals, high doses of Δ9-THC are neurotoxic in the hippocampus (Chan et al, 1998), where they decrease neuronal density (Landfield et al, 1988). Relative differences in the volume of a structure between groups may reflect reduced neuronal or glial density, cell size, or synaptic density (Yucel et al, 2008), and may derive from neurotoxicity, abnormal or attenuated neurogenesis, or premorbid (ie, before the onset of heavy cannabis use) differences. Reduced hippocampal volume may underlie a variety of symptoms of chronic heavy cannabis use, including deficits in verbal learning and spatial working memory (Ashtari et al, 2011; Rubino et al, 2009; Wise et al, 2009), while reduced amygdalar volume may be related to alterations in emotional processing among CU (Cornelius et al, 2010).
The interaction between rs202329 genotype and group on hippocampal volume indicates that the G allele of this SNP engendered smaller hippocampal volume only among CU; among HC, G-allele carriers had larger bilateral hippocampi than A-allele carriers. This result suggests the possibility of a gene by drug interaction, such that cannabis exposure may interact with CB1 receptors altered by rs2023239 in a manner that decreases the volume of brain structures in which these receptors are densely expressed. Ho and colleagues (Ho et al, 2011) recently reported a similar finding among individuals with schizophrenia, some of whom also had cannabis use disorders. They found that the presence of these disorders and variation at rs12720071, a SNP in the untranslated region of the fourth exon of CNR1, interacted in their effects on parietal white matter volume, such that rs12720071 genotype predicted smaller volume only among individuals who also had a cannabis use disorder. Given the fact that the rs2023239 G allele putatively increases CB1 expression in PFC (Hutchison et al, 2008), this SNP is a plausible candidate for a similar gene by drug interaction. However, it is also possible that smaller hippocampal volume and rs2023239 genotype independently preceded heavy cannabis use in the current study. Future longitudinal studies should determine whether volumetric differences are a precursor to or a consequence of heavy cannabis use.
In addition to our previous findings of association between rs202329 variance and cannabis withdrawal, craving, and cue-elicited brain activation (Filbey et al, 2010; Haughey et al, 2008), this SNP has been associated with other SUD intermediate phenotypes, including greater alcohol cue-elicited brain activation and greater subjective reward after consuming alcohol (Hutchison et al, 2008), greater alcohol craving at higher levels of alcohol consumption (van den Wildenberg et al, 2007), greater trait impulsivity (Ehlers et al, 2007), and, as part of a haplotype block with other CNR1 SNPs, greater severity of nicotine dependence (Chen et al, 2008). Although these other studies did not report participants’ cannabis use, it is highly comorbid with alcohol and nicotine use. Further, endocannabinoid signaling has widespread neural effects (Pazos et al, 2005), particularly with regard to addictive behavior (Onaivi, 2008). Thus, these associations are logical. The current study adds to these previous findings by raising the possibility, in light of the role of the hippocampus in addictive behavior, that reduced hippocampal volume among rs2023239 G-allele carriers may account for some variance in these associations. Future studies should further interrogate the effects of rs2023239 and other CNR1 SNPs, including CNR1 haplotypes, on brain volume and other CD intermediate phenotypes among cannabis users.
This study had several limitations. The HC sample, and particularly the subset of HC with an rs2023239 G allele (N=12), was relatively small, and outliers might have contributed to the magnitude of the genotype by group interactions. The associations reported here should be confirmed in larger samples, perhaps including individuals pre-screened on rs2023229 genotype to increase the number of G-allele carriers. Additionally, while the matched analyses compared age- and gender-matched groups, the larger CU sample was younger and more male than HC, reflecting the demographic characteristics of the broader CU population. Age and gender were covaried in analyses that compared HC to all CU, but analysis of covariance cannot entirely resolve systematic between-groups differences. However, given that women and older individuals have, on average, smaller subcortical volumes, the fact that volumes were instead smaller among younger and more male CU who were rs2023239 G-allele carriers supports the strength of the effect. Racial or ethnic differences between groups could also have affected results; though there was no evidence of allelic stratification by racial or ethnic group, genomic controls for race and ethnicity were not used. However, when continuous intermediate phenotypes, rather than dichotomous diagnostic phenotypes, are used as outcome measures in candidate genetic studies, population stratification may not represent a significant threat to validity (Hutchison et al, 2004). Nonetheless, these associations should be tested in larger samples of racially or ethnically homogenous groups. Further, CU had more alcohol and nicotine use than HC. While inclusion of these factors as covariates did not alter results, future research should use controls with more alcohol and nicotine use, as the significant prevalence of these behaviors among CU would likely preclude recruitment of heavy CU who do not use these substances. Because mood and anxiety disorders were not systematically diagnosed, these factors could also have confounded between-groups differences. Finally, while hippocampal and amygdalar volumes were computed automatically with FreeSurfer, rather than traced manually, FreeSurfer-computed hippocampal and amygdalar volumes correlate highly with manually traced volumes, with correlations for hippocampus in excess of 0.8 (Morey et al, 2009).
In summary, this study confirmed previous findings of association between heavy cannabis use and reduced hippocampal and amygdalar volumes, and suggested that reductions in hippocampal volume are moderated by a common variant in CNR1, indicating a gene by drug interaction on the volume of this structure. This association should be confirmed in an independent sample, and may merit consideration in future studies of the effects of heavy cannabis use on brain structure and function.
References
Agrawal A, Lynskey MT (2006). The genetic epidemiology of cannabis use, abuse and dependence. Addiction 101: 801–812.
Agrawal A, Lynskey MT (2009). Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction 104: 518–532.
Agrawal A, Wetherill L, Dick DM, Xuei X, Hinrichs A, Hesselbrock V et al (2009). Evidence for association between polymorphisms in the cannabinoid receptor 1 (CNR1) gene and cannabis dependence. Am J Med Genet B Neuropsychiatr Genet 150B: 736–740.
Ashtari M, Avants B, Cyckowski L, Cervellione KL, Roofeh D, Cook P et al (2011). Medial temporal structures and memory functions in adolescents with heavy cannabis use. J Psychiatr Res 45: 1055–1066.
Beck AT, Steer RA, Ball R, Ranieri W (1996). Comparison of beck depression inventories -IA and -II in psychiatric outpatients. J Pers Assess 67: 588–597.
Benegal V, Antony G, Venkatasubramanian G, Jayakumar PN (2007). Gray matter volume abnormalities and externalizing symptoms in subjects at high risk for alcohol dependence. Addict Biol 12: 122–132.
Benyamina A, Kebir O, Blecha L, Reynaud M, Krebs MO (2011). CNR1 gene polymorphisms in addictive disorders: a systematic review and a meta-analysis. Addict Biol 16: 1–6.
Bhattacharyya S, Morrison PD, Fusar-Poli P, Martin-Santos R, Borgwardt S, Winton-Brown T et al (2010). Opposite effects of delta-9-tetrahydrocannabinol and cannabidiol on human brain function and psychopathology. Neuropsychopharmacology 35: 764–774.
Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC et al (2004). A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. NeuroImage 23: 724–738.
Chan GC, Hinds TR, Impey S, Storm DR (1998). Hippocampal neurotoxicity of Delta9-tetrahydrocannabinol. J Neurosci 18: 5322–5332.
Cheer JF, Wassum KM, Heien ML, Phillips PE, Wightman RM (2004). Cannabinoids enhance subsecond dopamine release in the nucleus accumbens of awake rats. J Neurosci 24: 4393–4400.
Chen X, Williamson VS, An SS, Hettema JM, Aggen SH, Neale MC et al (2008). Cannabinoid receptor 1 gene association with nicotine dependence. Arch Gen Psychiatry 65: 816–824.
Cornelius JR, Aizenstein HJ, Hariri AR (2010). Amygdala reactivity is inversely related to level of cannabis use in individuals with comorbid cannabis dependence and major depression. Addict Behav 35: 644–646.
Cousijn J, Wiers RW, Ridderinkhof KR, van den Brink W, Veltman DJ, Goudriaan AE (2012). Grey matter alterations associated with cannabis use: results of a VBM study in heavy cannabis users and healthy controls. NeuroImage 59: 3845–3851.
Demirakca T, Sartorius A, Ende G, Meyer N, Welzel H, Skopp G et al (2011). Diminished gray matter in the hippocampus of cannabis users: possible protective effects of cannabidiol. Drug Alcohol Depend 114: 242–245.
Ehlers CL, Slutske WS, Lind PA, Wilhelmsen KC (2007). Association between single nucleotide polymorphisms in the cannabinoid receptor gene (CNR1) and impulsivity in southwest California Indians. Twin Res Hum Genet 10: 805–811.
Filbey FM, Schacht JP, Myers US, Chavez RS, Hutchison KE (2009). Marijuana craving in the brain. Proc Natl Acad Sci USA 106: 13016–13021.
Filbey FM, Schacht JP, Myers US, Chavez RS, Hutchison KE (2010). Individual and additive effects of the CNR1 and FAAH genes on brain response to marijuana cues. Neuropsychopharmacology 35: 967–975.
First MB, Spitzer RL, Gibbon M, Williams JBW (2002). Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Non-patient Edition. Biometrics Research, New York State Psychiatric Institute: New York.
Fischl B, Salat DH, van der Kouwe AJ, Makris N, Segonne F, Quinn BT et al (2004). Sequence-independent segmentation of magnetic resonance images. NeuroImage 23: S69–S84.
Gessa GL, Melis M, Muntoni AL, Diana M (1998). Cannabinoids activate mesolimbic dopamine neurons by an action on cannabinoid CB1 receptors. Eur J Pharmacol 341: 39–44.
Hartman CA, Hopfer CJ, Haberstick B, Rhee SH, Crowley TJ, Corley RP et al (2009). The association between cannabinoid receptor 1 gene (CNR1) and cannabis dependence symptoms in adolescents and young adults. Drug Alcohol Depend 104: 11–16.
Haughey HM, Marshall E, Schacht JP, Louis A, Hutchison KE (2008). Marijuana withdrawal and craving: influence of the cannabinoid receptor 1 (CNR1) and fatty acid amide hydrolase (FAAH) genes. Addiction 103: 1678–1686.
Herkenham M, Lynn AB, Johnson MR, Melvin LS, de Costa BR, Rice KC (1991). Characterization and localization of cannabinoid receptors in rat brain: a quantitative in vitro autoradiographic study. J Neurosci 11: 563–583.
Herman AI, Kranzler HR, Cubells JF, Gelernter J, Covault J (2006). Association study of the CNR1 gene exon 3 alternative promoter region polymorphisms and substance dependence. Am J Med Genet B Neuropsychiatr Genet 141B: 499–503.
Ho BC, Wassink TH, Ziebell S, Andreasen NC (2011). Cannabinoid receptor 1 gene polymorphisms and marijuana misuse interactions on white matter and cognitive deficits in schizophrenia. Schizophr Res 128: 66–75.
Hopfer CJ, Young SE, Purcell S, Crowley TJ, Stallings MC, Corley RP et al (2006). Cannabis receptor haplotype associated with fewer cannabis dependence symptoms in adolescents. Am J Med Genet B Neuropsychiatr Genet 141B: 895–901.
Hutchison KE, Haughey H, Niculescu M, Schacht J, Kaiser A, Stitzel J et al (2008). The incentive salience of alcohol: translating the effects of genetic variant in CNR1. Arch Gen Psychiatry 65: 841–850.
Hutchison KE, Stallings M, McGeary J, Bryan A (2004). Population stratification in the candidate gene study: fatal threat or red herring? Psychol Bull 130: 66–79.
Iversen L (2003). Cannabis and the brain. Brain 126: 1252–1270.
Landfield PW, Cadwallader LB, Vinsant S (1988). Quantitative changes in hippocampal structure following long-term exposure to delta 9-tetrahydrocannabinol: possible mediation by glucocorticoid systems. Brain Res 443: 47–62.
Lorenzetti V, Lubman DI, Whittle S, Solowij N, Yucel M (2010). Structural MRI findings in long-term cannabis users: what do we know? Subst Use Misuse 45: 1787–1808.
Matochik JA, Eldreth DA, Cadet JL, Bolla KI (2005). Altered brain tissue composition in heavy marijuana users. Drug Alcohol Depend 77: 23–30.
Morey RA, Petty CM, Xu Y, Hayes JP, Wagner II HR, Lewis DV et al (2009). A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. NeuroImage 45: 855–866.
Onaivi ES (2008). An endocannabinoid hypothesis of drug reward and drug addiction. Ann N Y Acad Sci 1139: 412–421.
Pazos MR, Nunez E, Benito C, Tolon RM, Romero J (2005). Functional neuroanatomy of the endocannabinoid system. Pharmacol Biochem Behav 81: 239–247.
Raudenbush SW, Bryk AS (2002). Hierarchical Linear Models: Applications and Data Analysis Methods, 2nd edn. Sage Publications: Thousand Oaks, CA.
Robbins TW, Ersche KD, Everitt BJ (2008). Drug addiction and the memory systems of the brain. Ann NY Acad Sci 1141: 1–21.
Rubino T, Realini N, Braida D, Guidi S, Capurro V, Vigano D et al (2009). Changes in hippocampal morphology and neuroplasticity induced by adolescent THC treatment are associated with cognitive impairment in adulthood. Hippocampus 19: 763–772.
Sobell LC, Sobell MB (1992). Timeline follow-back: a technique for assessing self-reported alcohol consumption. In: Allen JPL, R Z (eds). Measuring Alcohol Consumption: Psychosocial and Biochemical Methods. Humana Press: Totowa, NJ. pp 41–72.
Szabo B, Dorner L, Pfreundtner C, Norenberg W, Starke K (1998). Inhibition of GABAergic inhibitory postsynaptic currents by cannabinoids in rat corpus striatum. Neuroscience 85: 395–403.
Szabo B, Siemes S, Wallmichrath I (2002). Inhibition of GABAergic neurotransmission in the ventral tegmental area by cannabinoids. European J Neurosci 15: 2057–2061.
Takano Y, Tanaka T, Takano H, Hironaka N (2010). Hippocampal theta rhythm and drug-related reward-seekign behavior: an analysis of cocaine-induced conditioned place preference in rats. Brain Res 1342: 94–103.
van den Wildenberg E, Janssen RG, Hutchison KE, van Breukelen GJ, Wiers RW (2007). Polymorphisms of the dopamine D4 receptor gene (DRD4 VNTR) and cannabinoid CB1 receptor gene (CNR1) are not strongly related to cue-reactivity after alcohol exposure. Addict Biol 12: 210–220.
Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR (1999). Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect 107: 517–520.
Wechsler D (1999). Wechsler Abbreviated Scale of Intelligence (WASI). Harcourt Assessment: San Antonio, TX.
Wilson W, Mathew R, Turkington T, Hawk T, Coleman RE, Provenzale J (2000). Brain morphological changes and early marijuana use: a magnetic resonance and positron emission tomography study. J Addict Dis 19: 1–22.
Wise LE, Thorpe AJ, Lichtman AH (2009). Hippocampal CB(1) receptors mediate the memory impairing effects of Delta(9)-tetrahydrocannabinol. Neuropsychopharmacology 34: 2072–2080.
Xia Y, Portugal GS, Fakira AK, Melyan Z, Neve R, Lee HT et al (2011). Hippocampal GluA1-containing AMPA receptors mediate context-dependent sensitization to morphine. J Neurosci 31: 16279–16291.
Yucel M, Solowij N, Respondek C, Whittle S, Fornito A, Pantelis C et al (2008). Regional brain abnormalities associated with long-term heavy cannabis use. Arch Gen Psychiatry 65: 694–701.
Zhang PW, Ishiguro H, Ohtsuki T, Hess J, Carillo F, Walther D et al (2004). Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol Psychiatry 9: 916–931.
Acknowledgements
This work was supported by grants from NIDA (F31 DA021496 to JPS and K01 DA021632 to FMF) and NIAAA (T32 AA007474 to JPS). We gratefully acknowledge Diana South and Cathy Smith for their assistance during MRI data collection, Marilee Morgan for assistance with genotyping, and Patrick K Randall, PhD, for statistical consultation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Schacht, J., Hutchison, K. & Filbey, F. Associations between Cannabinoid Receptor-1 (CNR1) Variation and Hippocampus and Amygdala Volumes in Heavy Cannabis Users. Neuropsychopharmacol 37, 2368–2376 (2012). https://doi.org/10.1038/npp.2012.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2012.92
Keywords
This article is cited by
-
The cannabinoid receptor-1 gene interacts with stressful life events to increase the risk for problematic alcohol use
Scientific Reports (2022)
-
Genetic basis of cannabis use: a systematic review
BMC Medical Genomics (2021)
-
Sex and dependence related neuroanatomical differences in regular cannabis users: findings from the ENIGMA Addiction Working Group
Translational Psychiatry (2021)
-
Deconstructing the neurobiology of cannabis use disorder
Nature Neuroscience (2020)
-
BOLD responses to inhibition in cannabis-using adolescents and emerging adults after 2 weeks of monitored cannabis abstinence
Psychopharmacology (2020)