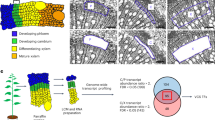
Abstract
Isolated protoplasts serve as a transient expression system that is highly representative of stable transgenics in terms of transcriptome responses. They can also be used as a cellular system to study gene transactivation and nucleocytoplasmic protein trafficking. They are particularly useful for systems studies in which stable transgenics and mutants are unavailable. We present a protocol for the isolation and transfection of protoplasts from wood-forming tissue, the stem-differentiating xylem (SDX), in the model woody plant Populus trichocarpa. The method involves tissue preparation, digestion of SDX cell walls, protoplast isolation and DNA transfection. Our approach is markedly faster and provides better yields than previous protocols; small (milligrams)- to large (20 g)-scale SDX preparations can be achieved in ∼60 s, with isolation of protoplasts and their subsequent transfection taking ∼50 min. Up to ten different samples can be processed simultaneously in this time scale. Our protocol gives a high yield (∼2.5 × 107 protoplasts per g of SDX) of protoplasts sharing 96% transcriptome identity with intact SDX.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sarkanen, K.V. Renewable resources for the production of fuels and chemicals. Science 191, 773–776 (1976).
Chiang, V. From rags to riches. Nat. Biotechnol. 20, 557–558 (2002).
Ragauskas, A. et al. The path forward for biofuels and biomaterials. Science 311, 484–489 (2006).
Hinchee, M. et al. Short-rotation woody crops for bioenergy and biofuels applications. In Vitro Cell. Dev. Bio. Plant 45, 619–629 (2009).
Evert, R.F. Esau's Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development (John Wiley & Sons, 2006).
Li, Q. et al. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 109, 14699–14704 (2012).
Lin, Y. et al. SND1 transcription factor-directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. Plant Cell 25, 4324–4341 (2013).
Lu, S. et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 110, 10848–10853 (2013).
Merkle, S.A. & Dean, J.F. Forest tree biotechnology. Curr. Opin. Biotechnol. 11, 298–302 (2000).
Song, J., Lu, S., Chen, Z., Lourenco, R. & Chiang, V.L. Genetic transformation of Populus trichocarpa genotype Nisqually-1: a functional genomic tool for woody plants. Plant Cell Physiol. 47, 1582–1589 (2006).
Chen, H. et al. Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 108, 21253–21258 (2011).
Leinhos, V. & Savidge, R.A. Isolation of protoplasts from developing xylem of Pinus banksiana and Pinus strobus. Can. J. For. Res. 23, 343–348 (1993).
Ahuja, M.R. Protoplast research in woody plants. Silvae Genet. 33, 32–37 (1984).
Teulieres, C., Grima-Pettenati, J., Curie, C., Teissie, J. & Boudet, A.M. Transient foreign gene expression in polyethylene/glycol treated or electropulsated Eucalyptus gunnii protoplasts. Plant Cell Tissue Organ Cult. 25, 125–132 (1991).
Manders, G., Dossantos, A.V.P., Vaz, F.B.D., Davey, M.R. & Power, J.B. Transient gene-expression in electroporated protoplasts of Eucalyptuscitriodora hook. Plant Cell Tissue Organ Cult. 30, 69–75 (1992).
Puite, K.J. Progress in plant protoplast research. Physiol. Plant. 85, 403–410 (1992).
Sun, J. et al. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 149, 1141–1153 (2009).
Tang, R. et al. The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 74, 367–380 (2010).
Gomez-Maldonado, J., Crespillo, R., Avila, C. & Canovas, F.M. Efficient preparation of maritime pine (Pinus pinaster) protoplasts suitable for transgene expression analysis. Plant Mol. Biol. Rep. 19, 361–366 (2001).
Harms, C.T. & Potrykus, I. Hormone-inhibition of Citrus protoplasts released by co-culturing with Nicotiana tabacum protoplasts–its significance for somatic hybrid selection. Plant Sci. Lett. 19, 295–301 (1980).
Cocking, E.C. Plant cell protoplasts–isolation and development. Annu. Rev. Plant. Physiol. 23, 29–50 (1972).
Yoo, S., Cho, Y. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Tan, B., Xu, M., Chen, Y. & Huang, M. Transient expression for functional gene analysis using Populus protoplasts. Plant Cell Tissue Organ Cult. 114, 11–18 (2013).
Wu, F. et al. Tape-Arabidopsis sandwich–a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009).
Guo, J. et al. Highly efficient isolation of Populus mesophyll protoplasts and its application in transient expression assays. PLoS ONE 7, e44908 (2012).
Kanwar, K., Bhardwaj, A. & Deepika, R. Efficient regeneration of plantlets from callus and mesophyll derived protoplasts of Robinia pseudoacacia L. Plant Cell Tissue Organ Cult. 96, 95–103 (2009).
Zhang, Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30 (2011).
Hong, S.Y., Seo, P.J., Cho, S.H. & Park, C.M. Preparation of leaf mesophyll protoplasts for transient gene expression in Brachypodium distachyon. J. Plant Biol. 55, 390–397 (2012).
Meyer, L., Serek, M. & Winkelmann, T. Protoplast isolation and plant regeneration of different genotypes of Petunia and Calibrachoa. Plant Cell Tissue Organ Cult. 99, 27–34 (2009).
Lung, S., Yanagisawa, M. & Chuong, S.D.X. Protoplast isolation and transient gene expression in the single-cell C4 species, Bienertia sinuspersici. Plant. Cell Rep. 30, 473–484 (2011).
Tuskan, G.A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 (2006).
Lockhart, J. Breaking down the complex regulatory web underlying lignin biosynthesis. Plant Cell 25, 4282 (2013).
Sheen, J. Signal transduction in Maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475 (2001).
Birnbaum, K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960 (2003).
Faraco, M., Sansebastiano, G.P.D., Spelt, K., Koes, R.E. & Quattrocchio, F.M. One protoplast is not the other. Plant Physiol. 156, 474–478 (2011).
Chupeau, M. et al. Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25, 2444–2463 (2013).
Fujii, H. et al. In vitro reconstitution of an abscisic acid signaling pathway. Nature 462, 660–664 (2009).
Nystedt, B. et al. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584 (2013).
Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013).
Courtois-Moreau, C.L. et al. A unique program for cell death in xylem fibers of Populus stem. Plant J. 58, 260–274 (2009).
Li, L. et al. The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13, 1567–1585 (2001).
Li, L. et al. Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. USA 100, 4939–4944 (2003).
Lu, S., Zhou, Y., Li, L. & Chiang, V.L. Distinct roles of cinnamate 4-hydroxylase genes in Populus. Plant Cell Physiol. 47, 905–914 (2006).
Lu, S., Li, L., Yi, X., Joshi, C.P. & Chiang, V.L. Differential expression of three eucalyptus secondary cell wall-related cellulose synthase genes in response to tension stress. J. Exp. Bot. 59, 681–695 (2008).
Li, J., Li, L. & Sheen, J. Protocol: a rapid and economical procedure for purification of plasmid or plant DNA with diverse applications in plant biology. Plant Methods 6, 1 (2010).
Miao, Y. & Jiang, L. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2, 2348–2353 (2007).
Nelson, B.K., Cai, X. & Nebenfuehr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007).
Sambrook, J. & Fritsch, E.F. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press (1989).
Schroeder, A. et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3 (2006).
Acknowledgements
This work was supported by the Office of Science (Biological and Environmental Research), US Department of Energy grant DE-SC000691 (to V.L.C.). We also thank the North Carolina State University Jordan Family Distinguished Professor Endowment for support.
Author information
Authors and Affiliations
Contributions
Y.-C.L., W.L., Q.L., Y.-H.S. and V.L.C. designed the experiment. Y.-C.L., W.L., H.C., Q.L., R.S., C.-Y.L., J.P.W., H.-C.C. and L.C. performed the experiments. Y.-C.L., W.L., Q.L., Y.-H.S. and V.L.C. analyzed the data. R.R.S., L.C. and G.-Z.Q. edited the manuscript. Y.-C.L., W.L. and V.L.C. wrote the paper. All authors discussed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Optimization of PEG-Mediated P. trichocarpa SDX Protoplast DNA Transfection (PDF 108 kb)
Footnotes: an represents the number of biological replicates of transfection efficiency test (see PROCEDURE) for each set of conditions. bThe optimized SDX protoplast transfection condition used for all PtrSND1-B1 overexpression experiments. Supplementary Table 1 was adapted from ref. 7 with permission from http://www.plantcell.org, Copyright American Society of Plant Biologists.
Rights and permissions
About this article
Cite this article
Lin, YC., Li, W., Chen, H. et al. A simple improved-throughput xylem protoplast system for studying wood formation. Nat Protoc 9, 2194–2205 (2014). https://doi.org/10.1038/nprot.2014.147
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.147
This article is cited by
-
Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa
Nature Plants (2023)
-
Protoplast isolation and transcriptome analysis of developing xylem in Pinus massoniana (Pinaceae)
Molecular Biology Reports (2022)
-
Transcriptional landscape of highly lignified poplar stems at single-cell resolution
Genome Biology (2021)
-
Qu-2, a robust poplar suspension cell line for molecular biology
Journal of Forestry Research (2021)
-
Simple protoplast isolation system for gene expression and protein interaction studies in pineapple (Ananas comosus L.)
Plant Methods (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.