Abstract
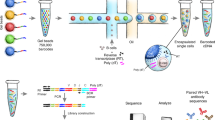
Methods to identify genes encoding immunoglobulin heavy and light chains from single B lymphocytes vary in efficiency, error rate and practicability. Here we describe a protocol to sequence and clone the variable antibody region of single antigen-specific mouse memory B cells for antibody production. After purification, antigen-specific mouse memory B cells are first single-cell-sorted by fluorescence-activated cell sorting (FACS), and V(D)J transcripts are amplified by RT-PCR. Fragments are then combined with linearized expression vectors, assembled in vitro as part of a sequence- and ligation-independent cloning (SLIC) reaction and then transformed into Escherichia coli. Purified vectors can then be used to produce monoclonal antibodies in HEK293E suspension cells. This protocol improves the amplification efficiency of antibody variable genes and accelerates the cloning workflow. Antibody sequences will be available in 3–4 d, and microgram to milligram amounts of antibodies are produced within 14 d. The new protocol should be useful for addressing fundamental questions about antigen-specific memory B cell responses, as well as for characterizing antigen-specific antibodies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Köhler, G. & Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 (1975).
Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Sastry, L. et al. Cloning of the immunological repertoire in Escherichia coli for generation of monoclonal catalytic antibodies: construction of a heavy chain variable region-specific cDNA library. Proc. Natl. Acad. Sci. USA 86, 5728–5732 (1989).
Huse, W.D. et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 246, 1275–1281 (1989).
Monaco, C., Nanchahal, J., Taylor, P. & Feldmann, M. Anti-TNF therapy: past, present and future. Int. Immunol. 27, 55–62 (2014).
Tiller, T., Busse, C.E. & Wardemann, H. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods 350, 183–193 (2009).
Kantor, A.B., Merrill, C.E., MacKenzie, J.D., Herzenberg, L.A. & Hillson, J.L. Development of the antibody repertoire as revealed by single-cell PCR of FACS-sorted B-cell subsets. Ann. NY Acad. Sci. 764, 224–227 (1995).
Max, E.E., Maizel, J.V. & Leder, P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J. Biol. Chem. 256, 5116–5120 (1981).
Johnston, C.M., Wood, A.L., Bolland, D.J. & Corcoran, A.E. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J. Immunol. 176, 4221–4234 (2006).
Brekke, K.M. & Garrard, W.T. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics 56, 490–505 (2004).
Thiebe, R. et al. The variable genes and gene families of the mouse immunoglobulin κ locus. Eur. J. Immunol. 29, 2072–2081 (1999).
Weiss, S. & Wu, G.E. Somatic point mutations in unrearranged immunoglobulin gene segments encoding the variable region of lambda light chains. EMBO J. 6, 927–932 (1987).
Wang, Z. et al. Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J. Immunol. Methods 233, 167–177 (2000).
Ehlers, M., Fukuyama, H., McGaha, T.L., Aderem, A. & Ravetch, J.V. TLR9/MyD88 signaling is required for class switching to pathogenic IgG2a and 2b autoantibodies in SLE. J. Exp. Med. 203, 553–561 (2006).
Casellas, R. et al. Ig allelic inclusion is a consequence of receptor editing. J. Exp. Med. 204, 153–160 (2007).
Sanchez, P., Marche, P.N., Rueff-Juy, D. & Cazenave, P.A. Mouse V lambda x gene sequence generates no junctional diversity and is conserved in mammalian species. J. Immunol. 144, 2816–2820 (1990).
Heinrichs, A., MILSTEIN, C. & Gherardi, E. Universal cloning and direct sequencing of rearranged antibody V genes using C region primers, biotin-captured cDNA and one-side PCR. J. Immunol. Methods 178, 241–251 (1995).
Ozawa, T., Kishi, H. & Muraguchi, A. Amplification and analysis of cDNA generated from a single cell by 5′-RACE: application to isolation of antibody heavy and light chain variable gene sequences from single B cells. BioTechniques 40, 469–470 (2006).
Aoki-Ota, M., Torkamani, A., Ota, T., Schork, N. & Nemazee, D. Skewed primary Ig repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J. Immunol. 188, 2305–2315 (2012).
Liang, J. et al. Development of the expressed immunoglobulin μ chain repertoire during maturation of mice B cells. Front. Agr. Sci. Eng. 1, 201–13 (2014).
Busse, C.E., Czogiel, I., Braun, P., Arndt, P.F. & Wardemann, H. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur. J. Immunol. 44, 597–603 (2013).
DeKosky, B.J. et al. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 21, 86–91 (2014).
Georgiou, G. et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 32, 158–168 (2014).
Smith, K. et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4, 372–384 (2009).
Tiller, T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124 (2008).
Wrammert, J. et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 (2008).
Dodev, T.S. et al. A tool kit for rapid cloning and expression of recombinant antibodies. Sci. Rep. 4, 1–10 (2014).
Li, M.Z. & Elledge, S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007).
Gitlin, A.D. et al. Independent roles of switching and hypermutation in the development and persistence of B lymphocyte memory. Immunity 44, 1–14 (2016).
Dosenovic, P. et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161, 1505–1515 (2015).
Scheid, J.F. et al. A method for identification of HIV gp140 binding memory B cells in human blood. J. Immunol. Methods 343, 65–67 (2009).
Soldemo, M., Pedersen, G. & Karlsson Hedestam, G. HIV-1 Env-specific memory and germinal center B cells in C57BL/6 mice. Viruses 6, 3400–3414 (2014).
Popp, M.W.-L., Antos, J.M. & Ploegh, H.L. Site-Specific Protein Labeling via Sortase-Mediated Transpeptidation 1–9 (John Wiley & Sons, Inc., 2001).
Improving Bioscience Research Reporting. The ARRIVE Guidelines for Reporting Animal Research 1–5 (2010).
Acknowledgements
We thank K. Velinzon and N. Thomas for assistance with single-cell FACS and Z. Jankovic for laboratory support. This work was supported by grants to M.C.N. from US National Institutes of Health (NIH) Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) UM1 AI100663, Bill and Melinda Gates Foundation (grant OPP1033115), NIH (grants AI037526 and U19 AI109632) and HIVRAD/P01 AI100148. L.v.B. is supported by grant no. UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), NIH Clinical and Translational Science Award (CTSA) program. A.D.G. is supported by MSTP grant T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. M.C.N. is a Howard Hughes Medical Institute Investigator. We thank members of the Nussenzweig and Ravetch laboratory for critical reading and helpful comments on the manuscript.
Author information
Authors and Affiliations
Contributions
L.v.B. designed the experiments; L.v.B. and M.C.N. wrote the manuscript; L.v.B., C.L. and S.A. performed the experiments; A.D.G., Q.W., A.G. and all other authors critically read the manuscript and contributed to the manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Agarose gel electrophoresis of single cell sorted IgG1+ PE specific memory B cells amplified with gamma chain specific primers
Cells were reverse transcribed with random primers. The 1st PCR was performed with the primers 1mFH_I – 1mFH_XI and 1mRG, primers for the nested sequencing PCR are indicated in Table 1. First two rows are from plate 1 with 51% (49/56 positive wells) amplification efficiency, rows 3 and 4 are from plate 2 with 59% (57/96 positive wells) amplification efficiency. Experiments were performed according to the protocols approved by the IACUC at Rockefeller University.
Supplementary information
Supplementary Table 1
Primers for first and sequencing PCR. (XLSX 32 kb)
Supplementary Table 2
Primers for cloning PCR. (PDF 215 kb)
Supplementary text and figures
Supplementary Figure 1. (PDF 238 kb)
Rights and permissions
About this article
Cite this article
von Boehmer, L., Liu, C., Ackerman, S. et al. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc 11, 1908–1923 (2016). https://doi.org/10.1038/nprot.2016.102
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.102
This article is cited by
-
Development and application of classical swine fever virus monoclonal antibodies derived from single B cells
Veterinary Research (2023)
-
CD47 masks pro-phagocytic ligands in cis on tumor cells to suppress antitumor immunity
Nature Immunology (2023)
-
Autoantibodies against chemokines post-SARS-CoV-2 infection correlate with disease course
Nature Immunology (2023)
-
Employment of a high throughput functional assay to define the critical factors that influence vaccine induced cross-variant neutralizing antibodies for SARS-CoV-2
Scientific Reports (2023)
-
Dynamics and durability of HIV-1 neutralization are determined by viral replication
Nature Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.