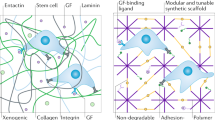
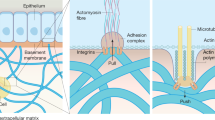
Abstract
2D surfaces offer simple analysis of cells in culture, yet these often yield different cell morphologies and responses from those observed in vivo. Considerable effort has therefore been expended on the generation of more tissue-like environments for the study of cell behavior in vitro. Purified matrix proteins provide a 3D scaffold that better mimics the in vivo situation; however, these are far removed from the complex tissue composition seen in vivo. Cell-derived matrices (CDMs) offer a more physiologically relevant alternative for studying in vivo-like cell behavior in vitro. In the protocol described here, fibroblasts cultured on gelatin-coated surfaces are maintained in the presence of ascorbic acid to strengthen matrix deposition over 1–3 weeks. The resulting fibrillar CDMs are denuded of cells, and other cells are subsequently cultured on them, after which their behavior is monitored. We also demonstrate how to use CDMs as an in vivo-relevant reductionist model for studying tumor-stroma-induced changes in carcinoma cell proliferation and migration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Engler, A.J., Sen, S., Sweeney, H.L. & Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Kai, F., Laklai, H. & Weaver, V.M. Force matters: biomechanical regulation of cell invasion and migration in disease. Trends Cell Biol. 26, 486–497 (2016).
Hirata, E. et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell 27, 574–588 (2015).
Nguyen-Ngoc, K.V. & Ewald, A.J. Mammary ductal elongation and myoepithelial migration are regulated by the composition of the extracellular matrix. J. Microsc. 251, 212–223 (2013).
Nguyen-Ngoc, K. et al. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods Mol. Biol. 1189, 135–162 (2015).
Caswell, P.T. et al. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell 13, 496–510 (2007).
Caswell, P.T. et al. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 183, 143–155 (2008).
Naba, A. et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell Proteomics 11, M111.014647 (2012).
Naba, A. et al. The extracellular matrix: tools and insights for the 'omics' era. Matrix Biol. 49, 10–24 (2016).
Franco-Barraza, J. et al. Matrix-regulated integrin αvβ5 maintains α5β1-dependent desmoplastic traits prognostic of neoplastic recurrence. Elife 6, e20600 (2017).
Kaukonen, R. et al. Normal stroma suppresses cancer cell proliferation via mechanosensitive regulation of JMJD1a-mediated transcription. Nat. Commun. 7, 12237 (2016).
Byron, A. et al. Glomerular cell cross-talk influences composition and assembly of extracellular matrix. J. Am. Soc. Nephrol. 25, 953–966 (2014).
Peuhu, E. et al. SHARPIN regulates collagen architecture and ductal outgrowth in the developing mouse mammary gland. EMBO J. 36, 165–182 (2017).
Rashid, S.T. et al. Proteomic analysis of extracellular matrix from the hepatic stellate cell line LX-2 identifies CYR61 and Wnt-5a as novel constituents of fibrotic liver. J. Proteome Res. 11, 4052–4064 (2012).
Soteriou, D. et al. Comparative proteomic analysis of supportive and unsupportive extracellular matrix substrates for human embryonic stem cell maintenance. J. Biol. Chem. 288, 18716–18731 (2013).
Amatangelo, M.D., Bassi, D.E., Klein-Szanto, A.J.P. & Cukierman, E. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am. J. Pathol. 167, 475–488 (2005).
Tello, M. et al. Generating and characterizing the mechanical properties of cell-derived matrices using atomic force microscopy. Methods 94, 85–100 (2016).
Ahlfors, J.W. & Billiar, K.L. Biomechanical and biochemical characteristics of a human fibroblast-produced and remodeled matrix. Biomaterials 28, 2183–2191 (2007).
Soucy, P.A., Werbin, J., Heinz, W., Hoh, J.H. & Romer, L.H. Microelastic properties of lung cell-derived extracellular matrix. Acta Biomater. 7, 96–105 (2011).
Jacquemet, G. et al. L-type calcium channels regulate filopodia stability and cancer cell invasion downstream of integrin signalling. Nat. Commun. 7, 13297 (2016).
Kaukonen, R. et al. Normal stroma suppresses cancer cell proliferation via mechanosensitive regulation of JMJD1a-mediated transcription. Nat. Commun. 7, 12237 (2016).
Jacquemet, G. et al. RCP-driven α5β1 recycling suppresses Rac and promotes RhoA activity via the RacGAP1-IQGAP1 complex. J. Cell Biol. 202, 917–935 (2013).
Jacquemet, G. et al. Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J. Cell. Sci. 126, 4121–4135 (2013).
Jacquemet, G. et al. FiloQuant reveals increased filopodia density during breast cancer progression. J. Cell Biol. pii: jcb.201704045 (2017).
Yamada, K.M. & Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601–610 (2007).
Cukierman, E., Pankov, R., Stevens, D.R. & Yamada, K.M. Taking cell-matrix adhesions to the third dimension. Science 294, 1708–1712 (2001).
Bass, M.D. et al. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J. Cell Biol. 177, 527–538 (2007).
Dozynkiewicz, M.A. et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell 22, 131–145 (2012).
Quiros, R.M. et al. Ovarian normal and tumor-associated fibroblasts retain in vivo stromal characteristics in a 3-D matrix-dependent manner. Gynecol. Oncol. 110, 99–109 (2008).
Grinnell, F., Fukamizu, H., Pawelek, P. & Nakagawa, S. Collagen processing, crosslinking, and fibril bundle assembly in matrix produced by fibroblasts in long-term cultures supplemented with ascorbic acid. Exp. Cell Res. 181, 483–491 (1989).
Wolf, K. et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069–1084 (2013).
Eyre, D.R., Paz, M.A. & Gallop, P.M. Cross-linking in collagen and elastin. Annu. Rev. Biochem. 53, 717–748 (1984).
Lelièvre, S., Weaver, V.M. & Bissell, M.J. Extracellular matrix signaling from the cellular membrane skeleton to the nuclear skeleton: a model of gene regulation. Recent Prog. Horm. Res. 51, 417–432 (1996).
Doyle, A.D., Carvajal, N., Jin, A., Matsumoto, K. & Yamada, K.M. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat. Commun. 6, 8720 (2015).
Petrie, R.J., Koo, H. & Yamada, K.M. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science 345, 1062–1065 (2014).
White, D.P., Caswell, P.T. & Norman, J.C. alpha v beta3 and alpha5beta1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J. Cell Biol. 177, 515–525 (2007).
Franco-Barraza, J., Beacham, D.A., Amatangelo, M.D. & Cukierman, E. Preparation of extracellular matrices produced by cultured and primary fibroblasts. Curr. Protoc. Cell Biol. 71, 10.9.34 (2016).
Gaudet, I.D. & Shreiber, D.I. Characterization of methacrylated type-I collagen as a dynamic, photoactive hydrogel. Biointerphases 7, 25 (2012).
Conklin, M.W. et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 (2011).
Pickup, M.W., Mouw, J.K. & Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15, 1243–1253 (2014).
Yun, S. et al. Interaction between integrin α5 and PDE4D regulates endothelial inflammatory signalling. Nat. Cell Biol. 18, 1043–1053 (2016).
Saneyasu, T., Akhtar, R. & Sakai, T. Molecular cues guiding matrix stiffness in liver fibrosis. Biomed. Res. Int. 2016, 2646212 (2016).
Boudaoud, A. et al. FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat. Protoc. 9, 457–463 (2014).
Lee, H. et al. FAP-overexpressing fibroblasts produce an extracellular matrix that enhances invasive velocity and directionality of pancreatic cancer cells. BMC Cancer 11, 245 (2011).
Kartasalo, K. et al. CytoSpectre: a tool for spectral analysis of oriented structures on cellular and subcellular levels. BMC Bioinformatics 16, 344 (2015).
Chaudhary, R. et al. Advanced quantitative imaging and biomechanical analyses of periosteal fibers in accelerated bone growth. Bone 92, 201–213 (2016).
Wells, R.G. The role of matrix stiffness in regulating cell behavior. Hepatology 47, 1394–1400 (2008).
Butcher, D.T., Alliston, T. & Weaver, V.M. A tense situation: forcing tumour progression. Nat. Rev. Cancer 9, 108–122 (2009).
Bomo, J. et al. Increasing 3D matrix rigidity strengthens proliferation and spheroid development of human liver cells in a constant growth factor environment. J. Cell Biochem. 117, 708–720 (2016).
Yeh, Y. et al. Matrix stiffness regulates endothelial cell proliferation through septin 9. PLoS ONE 7, e46889 (2012).
Muller, P.A.J. et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341 (2009).
Paul, N.R. et al. α5β1 integrin recycling promotes Arp2/3-independent cancer cell invasion via the formin FHOD3. J. Cell Biol. 210, 1013–1031 (2015).
Rainero, E. et al. Diacylglycerol kinase α controls RCP-dependent integrin trafficking to promote invasive migration. J. Cell Biol. 196, 277–295 (2012).
Petrie, R.J., Gavara, N., Chadwick, R.S. & Yamada, K.M. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197, 439–455 (2012).
Morgan, M.R. et al. Syndecan-4 phosphorylation is a control point for integrin recycling. Dev. Cell 24, 472–485 (2013).
Fitzpatrick, L.E. & McDevitt, T.C. Cell-derived matrices for tissue engineering and regenerative medicine applications. Biomater. Sci. 3, 12–24 (2015).
Kiss, A., Horvath, P., Rothballer, A., Kutay, U. & Csucs, G. Nuclear motility in glioma cells reveals a cell-line dependent role of various cytoskeletal components. PLoS ONE 9, e93431 (2014).
Paszek, M.J. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Acknowledgements
M. Georgiadou (University of Turku Centre for Biotechnology) is acknowledged for providing the bright-field images of CDMs presented in Figure 3. J. Norman (Beatson Institute for Cancer Research) is acknowledged for providing the TIFs. P. Caswell (University of Manchester) and J.F. Marshall (Barts Cancer Institute) are acknowledged for the A2780 and the MCF10 DCIS.COM cell lines, respectively. The Turku Centre for Biotechnology Imaging Core facility is acknowledged for help with the imaging. We gratefully acknowledge the following funding sources: Academy of Finland, European Research Council Consolidator Grant (no. 615258), the Sigrid Juselius Foundation and the Finnish Cancer Organization to J.I. R.K. was funded by the University of Turku/TUBS TuDMM doctoral program and G.J. was funded by an EMBO LTF.
Author information
Authors and Affiliations
Contributions
J.I., G.J., H.H. and R.K. organized and wrote the manuscript. H.H. provided the illustrated workflow and edited the manuscript. G.J. and R.K. produced the data. R.K. produced the video-guided material with help from G.J and J.I.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Workflow
This video outlines the entire workflow used to generate CDMs. (MP4 2139 kb)
Coverslip preparation
This video shows how to prepare gelatin-coated coverslips (Steps 1–6 of the protocol). (MP4 6555 kb)
Plating of fibroblasts
This video shows how to plate fibroblasts for CDM production (Steps 7–10 of the protocol). (MP4 21761 kb)
Extraction of CDMs.
This video shows how to extract CDMs (Steps 11–17 of the protocol). (MP4 21408 kb)
Staining of CDMs.
This video shows how to stain CDMs using immunofluorescence (Step 18A of the protocol). (MP4 8054 kb)
Analysis of cell proliferation.
This video shows how to analyze cell proliferation on CDMs using an IncuCyte Zoom live-cell incubator (Step 18B of the protocol). (MP4 7943 kb)
Quantification of cell migration.
This video shows how to quantify cell migration on CDMs using ImageJ and the Chemotaxis tool (Step 18C of the protocol). (MP4 4202 kb)
Migration of an ovarian carcinoma cell.
This video shows an ovarian carcinoma cell transiently expressing Lifeact RFP (to visualize the actin cytoskeleton) migrating on TIF CDMs (labeled in green using Alexa Fluor 488 recombinant fibronectin). Images were acquired on a spinning-disk microscope using a 63× objective. (MP4 4849 kb)
Rights and permissions
About this article
Cite this article
Kaukonen, R., Jacquemet, G., Hamidi, H. et al. Cell-derived matrices for studying cell proliferation and directional migration in a complex 3D microenvironment. Nat Protoc 12, 2376–2390 (2017). https://doi.org/10.1038/nprot.2017.107
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.107
This article is cited by
-
Content-aware frame interpolation (CAFI): deep learning-based temporal super-resolution for fast bioimaging
Nature Methods (2024)
-
Lung extracellular matrix modulates KRT5+ basal cell activity in pulmonary fibrosis
Nature Communications (2023)
-
In vitro spermatogenesis in artificial testis: current knowledge and clinical implications for male infertility
Cell and Tissue Research (2023)
-
Extracellular matrix derived from Wharton’s Jelly-derived mesenchymal stem cells promotes angiogenesis via integrin αVβ3/c-Myc/P300/VEGF
Stem Cell Research & Therapy (2022)
-
Recent advances in chemically defined and tunable hydrogel platforms for organoid culture
Bio-Design and Manufacturing (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.