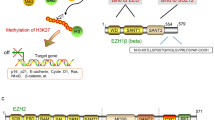
Abstract
Enhancer of zeste homologue 2 (EZH2), the catalytic subunit of Polycomb repressive complex 2 (PRC2), has attracted broad research attention in the past few years because of its involvement in the development and maintenance of many types of cancer and the use of specific EZH2 inhibitors in clinical trials. Several observations show that PRC2 can have both oncogenic and tumour-suppressive functions. We propose that these apparently opposing roles of PRC2 in cancer are a consequence of the molecular function of the complex in maintaining, rather than specifying, the transcriptional repression state of its several thousand target genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Helin, K. & Dhanak, D. Chromatin proteins and modifications as drug targets. Nature 502, 480–488 (2013).
Laugesen, A. & Helin, K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14, 735–751 (2014).
Simon, J. A. & Kingston, R. E. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 (2013).
Comet, I. & Helin, K. Revolution in the polycomb hierarchy. Nat. Struct. Mol. Biol. 21, 573–575 (2014).
Margueron, R. & Reinberg, D. The polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011).
Whitcomb, S. J., Basu, A., Allis, C. D. & Bernstein, E. Polycomb group proteins: an evolutionary perspective. Trends Genet. 23, 494–502 (2007).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Kleer, C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA 100, 11606–11611 (2003).
Bracken, A. P. et al. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22, 5323–5335 (2003).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 42, 181–185 (2010).
Sneeringer, C. J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA 107, 20980–20985 (2010).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 (2011).
Yap, D. B. et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2011).
Wigle, T. J. et al. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett. 585, 3011–3014 (2011).
McCabe, M. T. et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl Acad. Sci. USA 109, 2989–2994 (2012).
Majer, C. R. et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 586, 3448–3451 (2012).
Puda, A. et al. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am. J. Hematol. 87, 245–250 (2012).
Ntziachristos, P. et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 18, 298–301 (2012).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42, 722–726 (2010).
Khan, S. N. et al. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia 27, 1301–1309 (2013).
Nikoloski, G. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 42, 665–667 (2010).
Score, J. et al. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood 119, 1208–1213 (2012).
Ueda, T. et al. EED mutants impair polycomb repressive complex 2 in myelodysplastic syndrome and related neoplasms. Leukemia 26, 2557–2560 (2012).
De Raedt, T. et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514, 247–251 (2014).
Lee, W. et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 46, 1227–1232 (2014).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Sturm, D. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012).
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 (2012).
Knutson, S. K. et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl Acad. Sci. USA 110, 7922–7927 (2013).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Qi, W. et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl Acad. Sci. USA 109, 21360–21365 (2012).
Konze, K. D. et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem. Biol. 8, 1324–1334 (2013).
US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT01897571 (2013).
US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT02601937 (2015).
US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT02601950 (2015).
US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT02082977 (2014).
US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT02395601 (2015).
Mills, A. A. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat. Rev. Cancer 10, 669–682 (2010).
Koppens, M. & van Lohuizen, M. Context-dependent actions of Polycomb repressors in cancer. Oncogene 35, 1341–1352 (2016).
Blackledge, N. P., Rose, N. R. & Klose, R. J. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell. Biol. 16, 643–649 (2015).
Scelfo, A., Piunti, A. & Pasini, D. The controversial role of the Polycomb group proteins in transcription and cancer: how much do we not understand Polycomb proteins? FEBS J. 282, 1703–1722 (2015).
Lewis, E. B. A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978).
Jones, R. S. & Gelbart, W. M. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126, 185–199 (1990).
Simon, J., Chiang, A. & Bender, W. Ten different Polycomb group genes are required for spatial control of the AbdA and AbdB homeotic products. Development 114, 493–505 (1992).
Struhl, G. & Akam, M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 4, 3259–3264 (1985).
Akam, M. The molecular basis for metameric pattern in the Drosophila embryo. Development 101, 1–22 (1987).
Poux, S., McCabe, D. & Pirrotta, V. Recruitment of components of Polycomb group chromatin complexes in Drosophila. Development 128, 75–85 (2001).
Squazzo, S. L. et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16, 890–900 (2006).
Mohn, F. et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766 (2008).
Bracken, A. P., Dietrich, N., Pasini, D., Hansen, K. H. & Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 (2006).
Boyer, L. A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006).
Lee, T. I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006).
Nègre, N. et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 4, e170 (2006).
Schwartz, Y. B. et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38, 700–705 (2006).
Tolhuis, B. et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38, 694–699 (2006).
Abed, J. A. & Jones, R. S. H3K36me3 key to Polycomb-mediated gene silencing in lineage specification. Nat. Struct. Mol. Biol. 19, 1214–1215 (2012).
Beisel, C. & Paro, R. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 12, 123–135 (2011).
Schwartz, Y. B. & Pirrotta, V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 14, 853–864 (2013).
Voigt, P., Tee, W.-W. & Reinberg, D. A double take on bivalent promoters. Genes Dev. 27, 1318–1338 (2013).
Riising, E. M. et al. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 55, 347–360 (2014).
Yuan, W. et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337, 971–975 (2012).
Hosogane, M., Funayama, R., Nishida, Y., Nagashima, T. & Nakayama, K. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet. 9, e1003698 (2013).
Jermann, P., Hoerner, L., Burger, L. & Schubeler, D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl Acad. Sci. USA 111, E3415–E3421 (2014).
Mendenhall, E. M. et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 6, e1001244 (2010).
Klose, R. J., Cooper, S., Farcas, A. M., Blackledge, N. P. & Brockdorff, N. Chromatin sampling—an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 9, e1003717 (2013).
Ptashne, M. Epigenetics: core misconcept. Proc. Natl Acad. Sci. USA 110, 7101–7103 (2013).
Bödör, C. et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 122, 3165–3168 (2013).
Chang, C.-J. et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1–β-catenin signaling. Cancer Cell 19, 86–100 (2011).
Ciarapica, R. et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle 8, 172–175 (2009).
Collett, K. et al. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin. Cancer Res. 12, 1168–1174 (2006).
Gonzalez, M. E. et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc. Natl Acad. Sci. USA 111, 3098–3103 (2014).
Lohr, J. G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA 109, 3879–3884 (2012).
Lu, C. et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell 18, 185–197 (2010).
Okosun, J. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 (2014).
Puppe, J. et al. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to Polycomb repressive complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 11, R63 (2009).
Ryan, R. J. H. et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS ONE 6, e28585 (2011).
Takawa, M. et al. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci. 102, 1298–1305 (2011).
Varambally, S. et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 (2008).
Wagener, N. et al. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer 10, 524 (2010).
Wilson, B. G. et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010).
Yan, J. et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 121, 4512–4520 (2013).
Abdel-Wahab, O. et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 22, 180–193 (2012).
Amatangelo, M. D. et al. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell Cycle 12, 2113–2119 (2013).
Béguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Berg, T. et al. A transgenic mouse model demonstrating the oncogenic role of mutations in the polycomb-group gene EZH2 in lymphomagenesis. Blood 123, 3914–3924 (2014).
Cao, Q. et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 27, 7274–7284 (2008).
Herrera-Merchan, A. et al. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat. Commun. 3, 623 (2012).
Iliopoulos, D. et al. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 39, 761–772 (2010).
Kim, W. et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat. Chem. Biol. 9, 643–650 (2013).
Knutson, S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 8, 890–896 (2012).
Knutson, S. K. et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854 (2014).
Lane, A. a et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 46, 618–623 (2014).
Lee, S. C. W. et al. Polycomb repressive complex 2 (PRC2) suppresses Eμ-myc lymphoma. Blood 122, 2654–2663 (2013).
Lessard, J. et al. Functional antagonism of the Polycomb-group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 13, 2691–2703 (1999).
Min, J. et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-κB. Nat. Med. 16, 286–294 (2010).
Neff, T. et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl Acad. Sci. USA 109, 5028–5033 (2012).
Richie, E. R. et al. The Polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene 21, 299–306 (2002).
Sashida, G. et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat. Commun. 5, 4177 (2014).
Shi, J. et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene 32, 930–938 (2013).
Simon, C. et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 26, 651–656 (2012).
Tanaka, S. et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood 120, 1107–1117 (2012).
Tiwari, N. et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell 23, 768–783 (2013).
Wassef, M. et al. Impaired PRC2 activity promotes transcriptional instability and favors breast tumorigenesis. Genes Dev. 29, 2547–2562 (2015).
Abdel-Wahab, O. & Dey, A. The ASXL–BAP1 axis: new factors in myelopoiesis, cancer and epigenetics. Leukemia 27, 10–15 (2013).
Souroullas, G. P. et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 22, 632–640 (2016).
Kim, K. H. et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 21, 1491–1496 (2015).
Fillmore, C. M. et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 520, 239–242 (2015).
Serresi, M. et al. Polycomb repressive complex 2 is a barrier to KRAS-driven inflammation and epithelial–mesenchymal transition in non-small-cell lung cancer. Cancer Cell 29, 17–31 (2016).
Velichutina, I. et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 116, 5247–5255 (2010).
Ohm, J. E. et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237–242 (2007).
Schlesinger, Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39, 232–236 (2007).
Widschwendter, M. et al. Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158 (2007).
Easwaran, H. et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 22, 837–849 (2012).
Zhuang, J. et al. The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women's cancer. PLoS Genet. 8, e1002517 (2012).
Kia, S. K., Gorski, M. M., Giannakopoulos, S. & Verrijzer, C. P. SWI/SNF mediates Polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol. 28, 3457–3464 (2008).
Bracken, A. P. et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21, 525–530 (2007).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1–10 (2015).
Margueron, R. et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 (2008).
Pasini, D., Bracken, A. P., Jensen, M. R., Lazzerini Denchi, E. & Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 (2004).
Montgomery, N. D. et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 15, 942–947 (2005).
Ott, H. M. et al. A687V EZH2 is a driver of histone H3 lysine 27 (H3K27) hypertrimethylation. Mol. Cancer Ther. 13, 3062–3073 (2014).
Wu, H. et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS ONE 8, e83737 (2013).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013).
Bender, S. et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24, 660–672 (2013).
Chan, K.-M. et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013).
Acknowledgements
The authors thank members of the Helin laboratory for discussions. The work in the Helin laboratory is supported by The European Research Council (294666_DNAMET), the 7th framework programme of the European Union (4DCellFate and INGENIUM), the Danish Cancer Society, the Danish National Research Foundation (DNRF 82), the Danish Council for Strategic Research, the Danish Medical Research Council, the Novo Nordisk Foundation, The Lundbeck Foundation and through a centre grant from the Novo Nordisk Foundation (the Novo Nordisk Foundation Section for Stem Cell Biology in Human Disease).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Supplementary information
41568_2016_BFnrc201683_MOESM230_ESM.pdf
Supplementary information S1 (table) | Tumor sample analyses reveal that both gain and loss-offunction alterations of PRC2 occur in cancer (PDF 144 kb)
41568_2016_BFnrc201683_MOESM231_ESM.pdf
Supplementary information S2 (table) | Functional analyses reveal that PRC2 can play both oncogenic and tumour-suppressor roles (PDF 218 kb)
Rights and permissions
About this article
Cite this article
Comet, I., Riising, E., Leblanc, B. et al. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 16, 803–810 (2016). https://doi.org/10.1038/nrc.2016.83
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc.2016.83
This article is cited by
-
Bridging tissue repair and epithelial carcinogenesis: epigenetic memory and field cancerization
Cell Death & Differentiation (2024)
-
Regulation, functions and transmission of bivalent chromatin during mammalian development
Nature Reviews Molecular Cell Biology (2023)
-
Tumor-suppressive functions of protein lysine methyltransferases
Experimental & Molecular Medicine (2023)
-
Dissecting MENIN in bivalent gene regulation
Nature Cell Biology (2023)
-
HMGB1/SET/HAT1 complex-mediated SASH1 repression drives glycolysis and metastasis in lung adenocarcinoma
Oncogene (2023)