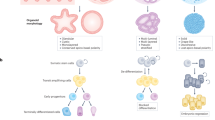
Key Points
-
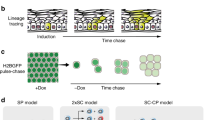
Lineage tracing in transgenic mice allows the study of stem, progenitor and cancer cells in their native environment and can give quantitative insight into cell behaviour. Experiments have two essential components: an inducible genetic switch, such as Cre recombinase, that activates the expression of the second essential component, the reporter gene, which allows the behaviour of cells to be followed.
-
Each lineage tracing system requires characterization to define the properties of the cell subpopulation being followed. Quantifying cell behaviour requires stringent controls and large-scale data sets.
-
Quantitative single cell lineage tracing has revealed cell behaviour in homeostatic epithelial tissues, such as the skin, oesophagus and intestine.
-
The squamous epithelia of the skin and oesophagus are maintained by a single progenitor cell population that generates progenitor and differentiated cells to achieve homeostasis. The epidermis contains quiescent stem cells but the oesophageal epithelium does not.
-
Intestinal epithelium is maintained by cycling stem cells expressing Lgr5 that reside in their niche at the base of the crypt. The restricted size of the niche is key in maintaining homeostasis.
-
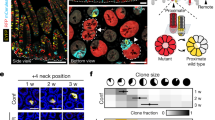
Oncogenes disrupt the balance between proliferation and differentiation in epithelial progenitors. For example, TP53-mutant cells in ultraviolet (UV)-exposed epidermis generate an excess of proliferating daughter cells, resulting in exponential expansion of mutant clones.
-
Preliminary studies in benign tumours reveal that intestinal adenomas are sustained by a population of Lgr5-expressing cells that have escaped the niche, and in epidermal papillomas stem and progenitor cells proliferate in a manner that is reminiscent of wound healing.
-
Lineage tracing in malignant lesions is more challenging owing to heterogeneity within and between tumours, but has already revealed unexpected early invasion and metastasis in pancreatic cancer models.
Abstract
For tumours to develop, mutations must disrupt tissue homeostasis in favour of deregulated proliferation. Genetic lineage tracing has uncovered the behaviour of proliferating cells that underpins the maintenance of epithelial tissues and the barriers that are broken in neoplastic transformation. In this Review, we focus on new insights revealed by quantifying the behaviour of normal, preneoplastic and tumour cells in epithelia in transgenic mice and consider their potential importance in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jones, P. H., Harper, S. & Watt, F. M. Stem cell patterning and fate in human epidermis. Cell 80, 83–93 (1995).
Li, A., Simmons, P. J. & Kaur, P. Identification and isolation of candidate human keratinocyte stem cells based on cell surface phenotype. Proc. Natl Acad. Sci. USA 95, 3902–3907 (1998).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 21, 70–71 (1999).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 166, 4 (2001).
Hoess, R. H. & Abremski, K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J. Mol. Biol. 181, 351–362 (1985).
Nagy, A. Cre recombinase: the universal reagent for genome tailoring. Genesis 26, 99–109 (2000).
Badea, T. C. et al. New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS ONE 466, e7859 (2009).
Ireland, H. et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of β-catenin. Gastroenterology 126, 1236–1246 (2004).
Kemp, R. et al. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res. 3266, e92 (2004).
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427–1429 (1995).
Littlewood, T. D., Hancock, D. C., Danielian, P. S., Parker, M. G. & Evan, G. I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23, 1686–1690 (1995).
Vasioukhin, V., Degenstein, L., Wise, B. & Fuchs, E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl Acad. Sci. USA 96, 8551–8556 (1999).
Hirrlinger, J. et al. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS ONE 466, e4286 (2009).
Beckervordersandforth, R. et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 7, 744–758 (2010).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). A multicolour confetti mouse enables quantitative lineage analysis in the intestine, revealing the role of restricted niche size in maintaining the epithelium.
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Murray, S. A., Eppig, J. T., Smedley, D., Simpson, E. M. & Rosenthal, N. Beyond knockouts: cre resources for conditional mutagenesis. Mamm. Genome 23, 587–599 (2012).
Clayton, E. et al. A single type of progenitor cell maintains normal epidermis. Nature 446, 185–189 (2007). The first study to use large-scale lineage tracing to quantify cell behaviour in vivo , revealing a new paradigm of tissue homeostasis.
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). Targeting an inducible Cre to a WNT target gene reveals that intestinal epithelium is maintained by stem cells at the base of the crypt.
Levy, V., Lindon, C., Harfe, B. D. & Morgan, B. A. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev. Cell 9, 855–861 (2005).
van Amerongen, R., Bowman, A. N. & Nusse, R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387–400 (2012).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Youssef, K. K. et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nature Cell Biol. 12, 299–305 (2010).
Heffner, C. S. et al. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nature Commun. 366, 1218 (2012).
Lapouge, G. et al. Identifying the cellular origin of squamous skin tumors. Proc. Natl Acad. Sci. USA 108, 7431–7436 (2011).
Mascre, G. et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489, 257–262 (2012).
Wong, V. W. et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nature Cell Biol. 14, 401–408 (2012).
Backman, C. M. et al. Generalized tetracycline induced Cre recombinase expression through the ROSA26 locus of recombinant mice. J. Neurosci. Methods 176, 16–23 (2009).
Loonstra, A. et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl Acad. Sci. USA 98, 9209–9214 (2001).
Zhu, J., Nguyen, M. T., Nakamura, E., Yang, J. & Mackem, S. Cre-mediated recombination can induce apoptosis in vivo by activating the p53 DNA damage-induced pathway. Genesis 50, 102–111 (2012).
Huh, W. J. et al. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology 142, 21–24.e27 (2012).
Lee, M. H. et al. Gene expression profiling of murine hepatic steatosis induced by tamoxifen. Toxicol. Lett. 199, 416–424 (2010).
Barker, N., van Oudenaarden, A. & Clevers, H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell 11, 452–460 (2012).
Tao, W. et al. Enhanced green fluorescent protein is a nearly ideal long-term expression tracer for hematopoietic stem cells, whereas DsRed-express fluorescent protein is not. Stem Cells 25, 670–678 (2007).
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnol. 22, 1567–1572 (2004).
Filonov, G. S. et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature Biotechnol. 29, 757–761 (2011).
Kretzschmar, K. & Watt, F. M. Lineage tracing. Cell 148, 33–45 (2012).
Morris, R. J. et al. Capturing and profiling adult hair follicle stem cells. Nature Biotechnol. 22, 411–417 (2004).
Ito, M. et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature Med. 11, 1351–1354 (2005).
Levy, V., Lindon, C., Zheng, Y., Harfe, B. D. & Morgan, B. A. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 21, 1358–1366 (2007).
Jensen, K. B. et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4, 427–439 (2009).
Snippert, H. J. et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 (2010).
Sangiorgi, E. & Capecchi, M. R. Bmi1 is expressed in vivo in intestinal stem cells. Nature Genet. 40, 915–920 (2008).
Snippert, H. J. & Clevers, H. Tracking adult stem cells. EMBO Rep. 12, 113–122 (2011).
Harfe, B. D. et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004).
Doupe, D. P., Klein, A. M., Simons, B. D. & Jones, P. H. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev. Cell 18, 317–323 (2010).
Klein, A. M., Doupe, D. P., Jones, P. H. & Simons, B. D. Kinetics of cell division in epidermal maintenance. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 7666, 021910 (2007).
Roshan, A. & Jones, P. H. Act your age: tuning cell behavior to tissue requirements in interfollicular epidermis. Semin. Cell Dev. Biol. 23, 884–889 (2012).
Jones, P. & Simons, B. D. Epidermal homeostasis: do committed progenitors work while stem cells sleep? Nature Rev. Mol. Cell Biol. 9, 82–88 (2008).
Marques-Pereira, J. P. & Leblond, C. P. Mitosis and differentiation in the stratified squamous epithelium of the rat esophagus. Am. J. Anat. 117, 73–87 (1965).
Goetsch, E. The structure of the mammalian esophagus. Am. J. Anat. 10, 1–39 (1910).
Doupe, D. P. et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337, 1091–1093 (2012). Lineage tracing and other approaches reveal that murine oesophagus has no quiescent stem cells, and is healed by the same progenitor cells that maintain the tissue.
Jones, P. H. Stem cell fate in proliferating tissues: equal odds in a game of chance. Dev. Cell 19, 489–490 (2010).
Takeda, N. et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011).
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
Powell, A. E. et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158 (2012).
Montgomery, R. K. et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl Acad. Sci. USA 108, 179–184 (2011).
Munoz, J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 31, 3079–3091 (2012).
van Es, J. H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature Cell Biol. 14, 1099–1104 (2012).
Slaughter, D. P., Southwick, H. W. & Smejkal, W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6, 963–968 (1953).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Jonason, A. S. et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc. Natl Acad. Sci. USA 93, 14025–14029 (1996).
Klein, A. M., Brash, D. E., Jones, P. H. & Simons, B. D. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc. Natl Acad. Sci. USA 107, 270–275 (2010). Quantitative analysis reveals that UV light drives the exponential expansion of p53-mutant clones in mouse and human epidermis.
Remenyik, E., Wikonkal, N. M., Zhang, W., Paliwal, V. & Brash, D. E. Antigen-specific immunity does not mediate acute regression of UVB-induced p53-mutant clones. Oncogene 22, 6369–6376 (2003).
Zhang, W., Remenyik, E., Zelterman, D., Brash, D. E. & Wikonkal, N. M. Escaping the stem cell compartment: sustained UVB exposure allows p53-mutant keratinocytes to colonize adjacent epidermal proliferating units without incurring additional mutations. Proc. Natl Acad. Sci. USA 98, 13948–13953 (2001).
Bennett, W. P. et al. p53 mutation and protein accumulation during multistage human esophageal carcinogenesis. Cancer Res. 52, 6092–6097 (1992).
Dainichi, T. et al. Chemical peeling by SA-PEG remodels photo-damaged skin: suppressing p53 expression and normalizing keratinocyte differentiation. J. Invest. Dermatol. 126, 416–421 (2006).
El-Domyati, M. M. et al. Effect of laser resurfacing on p53 expression in photoaged facial skin. Dermatol. Surg. 33, 668–675 (2007).
Cozzi, S. J. et al. Ingenol mebutate field-directed treatment of UVB-damaged skin reduces lesion formation and removes mutant p53 patches. J. Invest. Dermatol. 132, 1263–1271 (2011).
Rebel, H. G., Bodmann, C. A., van de Glind, G. C. & de Gruijl, F. R. UV-induced ablation of the epidermal basal layer including p53-mutant clones resets UV carcinogenesis showing squamous cell carcinomas to originate from interfollicular epidermis. Carcinogenesis 33, 714–720 (2012).
Driessens, G., Beck, B., Caauwe, A., Simons, B. D. & Blanpain, C. Defining the mode of tumour growth by clonal analysis. Nature 488, 527–530 (2012). Benign epidermal tumours result from the mobilization of stem- and progenitor-like cells, analogous to wound healing.
Schepers, A. G. et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735 (2012). Multicolour lineage tracing reveals that adenoma growth is underpinned by Lgr5 -expressing cells that have escaped the confines of the niche.
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Fodde, R., Smits, R. & Clevers, H. APC, signal transduction and genetic instability in colorectal cancer. Nature Rev. Cancer 1, 55–67 (2001).
Nakanishi, Y. et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nature Genet. 45, 98–103 (2012).
Dvorak, H. F. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 315, 1650–1659 (1986).
Schafer, M. & Werner, S. Cancer as an overhealing wound: an old hypothesis revisited. Nature Rev. Mol. Cell Biol. 9, 628–638 (2008).
Arwert, E. N., Hoste, E. & Watt, F. M. Epithelial stem cells, wound healing and cancer. Nature Rev. Cancer 12, 170–180 (2012).
Merlos-Suarez, A. et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 (2011).
Vermeulen, L. et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nature Cell Biol. 12, 468–476 (2010).
Vermeulen, L. et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl Acad. Sci. USA 105, 13427–13432 (2008).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762–1772 (2011).
Grachtchouk, M. et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J. Clin. Invest. 121, 1768–1781 (2011).
Wang, G. Y., Wang, J., Mancianti, M. L. & Epstein, E. H. Jr. Basal cell carcinomas arise from hair follicle stem cells in Ptch1+/− mice. Cancer Cell 19, 114–124 (2011).
White, A. C. et al. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc. Natl Acad. Sci. USA 108, 7425–7430 (2011).
Youssef, K. K. et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nature Cell Biol. 14, 1282–1294 (2012).
Seykora, J. T. & Cotsarelis, G. Keratin 15-positive stem cells give rise to basal cell carcinomas in irradiated Ptch1+/− mice. Cancer Cell 19, 5–6 (2011).
Epstein, E. H. Jr. Mommy - where do tumors come from? J. Clin. Invest. 121, 1681–1683 (2011).
Rhim, A. D. et al. EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361 (2012). Lineage tracing in a pancreatic cancer model reveals that cells acquire an invasive and metastatic phenotype before detectable tumour formation.
Bayne, L. J. et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 21, 822–835 (2012).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011).
Merlo, L. M. et al. A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev. Res. 3, 1388–1397 (2010).
McCaughan, F. et al. Genomic evidence of pre-invasive clonal expansion, dispersal and progression in bronchial dysplasia. J. Pathol. 224, 153–159 (2011).
Klein, A. M. & Simons, B. D. Universal patterns of stem cell fate in cycling adult tissues. Development 138, 3103–3111 (2011).
Lopez-Garcia, C., Klein, A. M., Simons, B. D. & Winton, D. J. Intestinal stem cell replacement follows a neutral drift. Science 330, 822–825 (2010).
Simons, B. D. & Clevers, H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell 145, 851–862 (2011).
Acknowledgements
The authors thank D. Doupé, K. Jensen, A. Klein, K. Murai, A. Roshan, B. Simons and D.Winton for illuminating discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Alcolea, M., Jones, P. Tracking cells in their native habitat: lineage tracing in epithelial neoplasia. Nat Rev Cancer 13, 161–171 (2013). https://doi.org/10.1038/nrc3460
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3460
This article is cited by
-
The role of microRNAs in the modulation of cancer-associated fibroblasts activity during pancreatic cancer pathogenesis
Journal of Physiology and Biochemistry (2023)
-
Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives
Molecular Cancer (2021)
-
The promising role of noncoding RNAs in cancer-associated fibroblasts: an overview of current status and future perspectives
Journal of Hematology & Oncology (2020)
-
A single-progenitor model as the unifying paradigm of epidermal and esophageal epithelial maintenance in mice
Nature Communications (2020)
-
A framework for advancing our understanding of cancer-associated fibroblasts
Nature Reviews Cancer (2020)