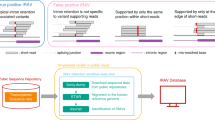
Abstract
When genome variants are identified in genomic DNA, especially during routine analysis of disease-associated genes, their functional implications might not be immediately evident. Distinguishing between a genomic variant that changes the phenotype and one that does not is a difficult task. An increasing amount of evidence indicates that genomic variants in both coding and non-coding sequences can have unexpected deleterious effects on the splicing of the gene transcript. So how can benign polymorphisms be distinguished from disease-associated splicing mutations?
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Thude, H., Hundrieser, J., Wonigeit, K. & Schwinzer, R. A point mutation in the human CD45 gene associated with defective splicing of exon A. Eur. J. Immunol. 25, 2101–2106 (1995).
Varani, L. et al. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl Acad. Sci. USA 96, 8229–8234 (1999).
Ars, E. et al. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 9, 237–247 (2000).
Teraoka, S. N. et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 64, 1617–1631 (1999).
Pagani, F. et al. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 12, 1111–1120 (2003).
Pagani, F. et al. A new type of mutation causes a splicing defect in ATM. Nature Genet. 30, 426–429 (2002).
Pagani, F., Buratti, E., Stuani, C. & Baralle, F. E. Missense, nonsense and neutral mutations define juxtaposed regulatory elements of splicing in CFTR exon 9. J. Biol. Chem. 278, 26580–26588 (2003).
Cartegni, L., Chew, S. L. & Krainer, A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 3, 285–298 (2002).
Kashima, T. & Manley, J. L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nature Genet. 34, 460–463 (2003).
Liu, H. X., Cartegni, L., Zhang, M. Q. & Krainer, A. R. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nature Genet. 27, 55–58 (2001).
Jiang, Z. et al. Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2 β. J. Biol. Chem. 278, 18997–19007 (2003).
Ishii, S., Nakao, S., Minamikawa-Tachino, R., Desnick, R. J. & Fan, J. Q. Alternative splicing in the α-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 70, 994–1002 (2002).
Faustino, N. A. & Cooper, T. A. Pre-mRNA splicing and human disease. Genes Dev. 17, 419–437 (2003).
Fernandez-Cadenas, I. et al. Splicing mosaic of the myophosphorylase gene due to a silent mutation in McArdle disease. Neurology 61, 1432–1434 (2003).
Black, D. L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 (2003).
Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Chillon, M. et al. Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. N. Engl. J. Med. 332, 1475–1480 (1995).
Chu, C. S., Trapnell, B. C., Curristin, S., Cutting, G. R. & Crystal, R. G. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nature Genet. 3, 151–156 (1993).
Larriba, S. et al. Testicular CFTR splice variants in patients with congenital absence of the vas deferens. Hum. Mol. Genet. 7, 1739–1743 (1998).
Mak, V., Jarvi, K. A., Zielenski, J., Durie, P. & Tsui, L. C. Higher proportion of intact exon 9 CFTR mRNA in nasal epithelium compared with vas deferens. Hum. Mol. Genet. 6, 2099–2107 (1997).
D'Souza, I. & Schellenberg, G. D. Determinants of 4-repeat tau expression. Coordination between enhancing and inhibitory splicing sequences for exon 10 inclusion. J. Biol. Chem. 275, 17700–17709 (2000).
Zhang, M. Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 7, 919–932 (1998).
Baralle, M. et al. Identification of a mutation that perturbs NF1 agene splicing using genomic DNA samples and a minigene assay. J. Med. Genet. 40, 220–222 (2003).
Roca, X., Sachidanandam, R. & Krainer, A. R. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 31, 6321–6333 (2003).
Matsushima, M. et al. Mutation analysis of the BRCA1 gene in 76 Japanese ovarian cancer patients: four germline mutations, but no evidence of somatic mutation. Hum. Mol. Genet. 4, 1953–1956 (1995).
McCullough, A. J. & Berget, S. M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 17, 4562–4571 (1997).
Forch, P. et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre- mRNA splicing. Mol. Cell 6, 1089–1098 (2000).
Metherell, L. A. et al. Pseudoexon activation as a novel mechanism for disease resulting in atypical growth-hormone insensitivity. Am. J. Hum. Genet. 69, 641–646 (2001).
Christie, P. T., Harding, B., Nesbit, M. A., Whyte, M. P. & Thakker, R. V. X-linked hypophosphatemia attributable to pseudoexons of the PHEX gene. J. Clin. Endocrinol. Metab. 86, 3840–3844 (2001).
Lev-Maor, G., Sorek, R., Shomron, N. & Ast, G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300, 1288–1291 (2003).
Buratti, E. et al. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 24, 1387–1400 (2004).
Muro, A. F. et al. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 19, 2657–2671 (1999).
Shen, L. X., Basilion, J. P. & Stanton, V. P. Jr. Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc. Natl Acad. Sci. USA 96, 7871–7876 (1999).
Nagel, R. J., Lancaster, A. M. & Zahler, A. M. Specific binding of an exonic splicing enhancer by the pre-mRNA splicing factor SRp55. RNA 4, 11–23 (1998).
Shi, H., Hoffman, B. E. & Lis, J. T. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 17, 2649–2657 (1997).
Damgaard, C. K., Tange, T. O. & Kjems, J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8, 1401–1415 (2002).
Grover, A. et al. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 274, 15134–15143 (1999).
Jiang, Z., Cote, J., Kwon, J. M., Goate, A. M. & Wu, J. Y. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 20, 4036–4048 (2000).
Buratti, E. et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20, 1774–1784 (2001).
Pagani, F. et al. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 275, 21041–21047 (2000).
Buratti, E., Brindisi, A., Pagani, F. & Baralle, F. E. Why does a variation in the number of TG repeats in CFTR intron 8 influence disease penetrance? Am. J. Hum. Genet. (in the press).
Buchner, D. A., Trudeau, M. & Meisler, M. H. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science 301, 967–969 (2003).
Nadeau, J. H. Genetics. Modifying the message. Science 301, 927–928 (2003).
Kearney, J. A. et al. Molecular and pathological effects of a modifier gene on deficiency of the sodium channel Scn8a (Nav1.6). Hum. Mol. Genet. 11, 2765–2775 (2002).
Johnson, J. M. et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144 (2003).
Maniatis, T. & Reed, R. An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002).
Proudfoot, N. J., Furger, A. & Dye, M. J. Integrating mRNA processing with transcription. Cell 108, 501–512 (2002).
Roberts, G. C., Gooding, C., Mak, H. Y., Proudfoot, N. J. & Smith, C. W. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26, 5568–5572 (1998).
Proudfoot, N. J. Dawdling polymerases allow introns time to splice. Nature Struct. Biol. 10, 876–878 (2003).
Attanasio, C., David, A. & Neerman-Arbez, M. Outcome of donor splice site mutations accounting for congenital afibrinogenemia reflects order of intron removal in the fibrinogen α gene (FGA). Blood 101, 1851–1856 (2003).
Schwarze, U., Starman, B. J. & Byers, P. H. Redefinition of exon 7 in the COL1A1 gene of type I collagen by an intron 8 splice-donor-site mutation in a form of osteogenesis imperfecta: influence of intron splice order on outcome of splice-site mutation. Am. J. Hum. Genet. 65, 336–344 (1999).
Hoogendoorn, B. et al. Functional analysis of human promoter polymorphisms. Hum. Mol. Genet. 12, 2249–2254 (2003).
Pagani, F., Stuani, C., Zuccato, E., Kornblihtt, A. R. & Baralle, F. E. Promoter architecture modulates CFTR exon 9 skipping. J. Biol. Chem. 278, 1511–1517 (2003).
Cramer, P. et al. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4, 251–258 (1999).
Cramer, P., Pesce, C. G., Baralle, F. E. & Kornblihtt, A. R. Functional association between promoter structure and transcript alternative splicing. Proc. Natl Acad. Sci. USA 94, 11456–11460 (1997).
Monsalve, M. et al. Direct coupling of transcription and mRNA processing through the thermogenic co-activator PGC-1. Mol. Cell 6, 307–316 (2000).
Zhang, C. et al. Nuclear co-activator-62 kDa/Ski-interacting protein is a nuclear matrix-associated co-activator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278, 35325–35336 (2003).
Cargill, M. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 22, 231–238 (1999).
Halushka, M. K. et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature Genet. 22, 239–247 (1999).
Fairbrother, W. G., Yeh, R. F., Sharp, P. A. & Burge, C. B. Predictive identification of exonic splicing enhancers in human genes. Science 297, 1007–1013 (2002).
Cartegni, L. & Krainer, A. R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet. 30, 377–384 (2002).
Tian, M. & Maniatis, T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 74, 105–114 (1993).
Lorson, C. L. & Androphy, E. J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 9, 259–265 (2000).
Acknowledgements
This work was supported by the Telethon Onlus Foundation, Italy, the Fondo Investimenti per la Ricerca di Base and the Associazione Italiana per la Ricerca sul Cancro.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
LocusLink
OMIM
Swiss-Prot
FURTHER INFORMATION
Glossary
- ALTERNATIVELY SPLICED ISOFORMS
-
RNA isoforms that are generated by alternative use of splice sites, which leads to variation in which exons are included in the mRNA and subsequently translated.
- ATAXIA TELANGIECTASIA
-
An autosomal recessive disorder that involves cerebellar degeneration, immunodeficiency, chromosomal instability, radiosensitivity and cancer predisposition.
- COMPOSITE EXONIC REGULATORY ELEMENT OF SPLICING
-
(CERES). Short exonic RNA sequences (5–12 bases) that contain overlapping enhancer and silencer sequences. The presence of such elements is indicated when scanning mutagenesis analyses reveal that different mutations at nearby positions or even at the same position have opposite effects on splicing efficiency.
- CRYPTIC SPLICE SITES
-
Pseudo splice sites that are activated as a consequence of a mutation elsewhere in the gene.
- EXON SKIPPING
-
Exclusion of an exon that is normally included in the mRNA.
- EXON SPLICING ENHANCER AND EXON SPLICING SILENCER (ESE, ESS); INTRON SPLICING ENHANCER AND INTRON SPLICING SILENCER (ISE, ISS)
-
Sequences in the pre-mRNA that enhance or reduce the efficiency of splicing. In general, exonic enhancers or silencers are shorter (∼6 bases) than the intronic ones, which can be hundreds of bases long.
- HETEROGENOUS NUCLEAR RIBONUCLEOPROTEIN PARTICLES
-
(hnRNP). A class of diverse RNA-binding proteins that associate with nascent pre-mRNA.
- HYBRID MINIGENE
-
A simplified laboratory version of a natural gene that contains one of more of the gene's exons and introns.
- NEUROFIBROMATOSIS TYPE 1
-
An autosomal dominant disorder that is particularly characterized by cafe-au-lait spots and fibromatous tumours of the skin.
- PSEUDO EXON
-
A pre-mRNA sequence that resembles an exon, both in its size and the presence of flanking pseudo splice sites, but that the splicing machinery does not normally recognize.
- PSEUDO SPLICE SITES
-
Sequences that are identical to normal splice sites but that are not normally used in splicing.
- SIMPLE SEQUENCE REPEAT
-
A sequence that consists largely of a tandem repeat of a specific K-mer (such as (TG)11).
- SITE-DIRECTED MUTAGENESIS
-
A method that is used to substitute a specific nucleotide into a DNA sequence.
- TRANSESTERIFICATION
-
A reaction that breaks and makes chemical bonds (in this case, phosphodiester bonds) in a coordinated transfer so that energy is required.
Rights and permissions
About this article
Cite this article
Pagani, F., Baralle, F. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet 5, 389–396 (2004). https://doi.org/10.1038/nrg1327
Issue Date:
DOI: https://doi.org/10.1038/nrg1327
This article is cited by
-
Intronic position +9 and −9 are potentially splicing sites boundary from intronic variants analysis of whole exome sequencing data
BMC Medical Genomics (2023)
-
MC1R and age heteroclassification of face phenotypes in the Rio Grande do Sul population
International Journal of Legal Medicine (2023)
-
A Study of Combined Genotype Effects of SHCBP1 on Wool Quality Traits in Chinese Merino
Biochemical Genetics (2023)
-
Genomic architecture of phenotypic extremes in a wild cervid
BMC Genomics (2022)
-
A novel nonsense variant in ARID1B causing simultaneous RNA decay and exon skipping is associated with Coffin-Siris syndrome
Human Genome Variation (2022)