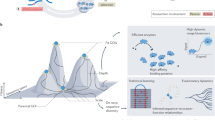
Key Points
-
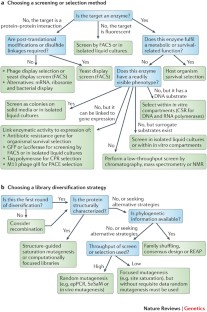
Directed evolution is a cyclic process that alternates between gene diversification and screening for or selection of functional gene variants.
-
Library size limitations can be overcome by focusing library diversity on residues implicated by molecular structures, computational models or phylogenetic data. In cases in which there is limited information, random mutagenesis can be used to interrogate the uncertain determinants of protein function.
-
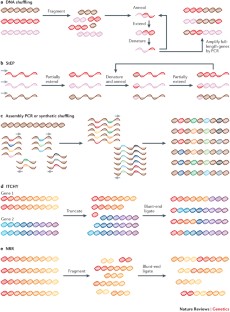
Recombination methodologies access new combinations of functional variation and can shuffle disparate genetic elements to yield new chimeric proteins.
-
Low-throughput screens can directly measure individual phenotypes and thus accurately isolate desired subpopulations. Screen throughput can be increased using indirect visible reporters that are strongly coupled to the desired phenotypes.
-
Selections isolate functional variants through selective replication schemes or physical segregation. Selections operate simultaneously on entire populations and thus offer unparalleled throughput.
Abstract
Directed evolution has proved to be an effective strategy for improving or altering the activity of biomolecules for industrial, research and therapeutic applications. The evolution of proteins in the laboratory requires methods for generating genetic diversity and for identifying protein variants with desired properties. This Review describes some of the tools used to diversify genes, as well as informative examples of screening and selection methods that identify or isolate evolved proteins. We highlight recent cases in which directed evolution generated enzymatic activities and substrate specificities not known to exist in nature.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wright, S. I. et al. The effects of artificial selection on the maize genome. Science 308, 1310–1314 (2005).
Driscoll, C. A., Macdonald, D. W. & O'Brien, S. J. From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl Acad. Sci. USA 106 (Suppl. 1), 9971–9978 (2009).
Umeno, D., Tobias, A. V. & Arnold, F. H. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 69, 51–78 (2005).
Atsumi, S. & Liao, J. C. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl. Environ. Microbiol. 74, 7802–7808 (2008).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Zhang, Y. X. et al. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415, 644–646 (2002).
Alper, H., Moxley, J., Nevoigt, E., Fink, G. R. & Stephanopoulos, G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314, 1565–1568 (2006).
Coelho, P. S., Brustad, E. M., Kannan, A. & Arnold, F. H. Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes. Science 339, 307–310 (2013).
McIsaac, R. S. et al. Directed evolution of a far-red fluorescent rhodopsin. Proc. Natl Acad. Sci. USA 111, 13034–13039 (2014).
Jespers, L. S., Roberts, A., Mahler, S. M., Winter, G. & Hoogenboom, H. R. Guiding the selection of human-antibodies from phage display repertoires to a single epitope of an antigen. Biotechnology 12, 899–903 (1994).
Lai, Y. P., Huang, J., Wang, L. F., Li, J. & Wu, Z. R. A new approach to random mutagenesis in vitro. Biotechnol. Bioeng. 86, 622–627 (2004).
Myers, R. M., Lerman, L. S. & Maniatis, T. A general method for saturation mutagenesis of cloned DNA fragments. Science 229, 242–247 (1985).
Freese, E. Specific mutagenic effect of base analogues on Phage-T4. J. Mol. Biol. 1, 87–105 (1959).
Bridges, B. A. & Woodgate, R. Mutagenic repair in Escherichia coli: products of the recA gene and of the umuD and umuC genes act at different steps in UV-induced mutagenesis. Proc. Natl Acad. Sci. USA 82, 4193–4197 (1985).
Cox, E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu. Rev. Genet. 10, 135–156 (1976).
Greener, A., Callahan, M. & Jerpseth, B. An efficient random mutagenesis technique using an E. coli mutator strain. Mol. Biotechnol. 7, 189–195 (1997).
Scheuermann, R., Tam, S., Burgers, P. M. J., Lu, C. & Echols, H. Identification of the ε-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc. Natl Acad. Sci. USA 80, 7085–7089 (1983).
Ravikumar, A., Arrieta, A. & Liu, C. C. An orthogonal DNA replication system in yeast. Nat. Chem. Biol. 10, 175–177 (2014).
Leung, D. W., Chen, E. & Goeddel, D. V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1, 11–15 (1989).
Zaccolo, M., Williams, D. M., Brown, D. M. & Gherardi, E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 255, 589–603 (1996).
Eckert, K. A. & Kunkel, T. A. High fidelity DNA synthesis by the Thermus Aquaticus DNA polymerase. Nucleic Acids Res. 18, 3739–3744 (1990).
Gupta, R. D. & Tawfik, D. S. Directed enzyme evolution via small and effective neutral drift libraries. Nat. Methods 5, 939–942 (2008).
Cadwell, R. C. & Joyce, G. F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2, 28–33 (1992). This seminal study in optimizing the conditions for epPCR is a must-read for all scientists performing random mutagenesis.
Vanhercke, T., Ampe, C., Tirry, L. & Denolf, P. Reducing mutational bias in random protein libraries. Anal. Biochem. 339, 9–14 (2005).
Wong, T. S., Tee, K. L., Hauer, B. & Schwaneberg, U. Sequence saturation mutagenesis (SeSaM): a novel method for directed evolution. Nucleic Acids Res. 32, e26 (2004).
Wells, J. A., Vasser, M. & Powers, D. B. Cassette mutagenesis: an efficient method for generation of multiple mutations at defined sites. Gene 34, 315–323 (1985).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–341 (2009).
Quan, J. Y. & Tian, J. D. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4, e6441 (2009).
Nour-Eldin, H. H., Geu-Flores, F. & Halkier, B. A. User cloning and user fusion: the ideal cloning techniques for small and big laboratories. Methods Mol. Biol. 643, 185–200 (2010).
Reidhaarolson, J. F. & Sauer, R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science 241, 53–57 (1988).
Lehmann, M., Pasamontes, L., Lassen, S. F. & Wyss, M. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta 1543, 408–415 (2000).
Chen, F. et al. Reconstructed evolutionary adaptive paths give polymerases accepting reversible terminators for sequencing and SNP detection. Proc. Natl Acad. Sci. USA 107, 1948–1953 (2010).
Cherny, I. et al. Engineering V-type nerve agents detoxifying enzymes using computationally focused libraries. ACS Chem. Biol. 8, 2394–2403 (2013). This paper nicely demonstrates how computational modelling can identify beneficial mutations, which can be stochastically incorporated into gene libraries.
Das, R. & Baker, D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 77, 363–382 (2008).
Wijma, H. J. et al. Computationally designed libraries for rapid enzyme stabilization. Protein Eng. Des. Sel. 27, 49–58 (2014).
Herman, A. & Tawfik, D. S. Incorporating synthetic oligonucleotides via gene reassembly (ISOR): a versatile tool for generating targeted libraries. Protein Eng. Des. Sel. 20, 219–226 (2007).
Stemmer, W. P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994). This study is the first to establish a method for homologous recombination of evolving protein populations.
Coco, W. M. et al. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat. Biotechnol. 19, 354–359 (2001).
Muller, K. M. et al. Nucleotide exchange and excision technology (NExT) DNA shuffling: a robust method for DNA fragmentation and directed evolution. Nucleic Acids Res. 33, e117 (2005).
Zhao, H., Giver, L., Shao, Z., Affholter, J. A. & Arnold, F. H. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 16, 258–261 (1998).
Stemmer, W. P., Crameri, A., Ha, K. D., Brennan, T. M. & Heyneker, H. L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164, 49–53 (1995).
Ness, J. E. et al. Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat. Biotechnol. 20, 1251–1255 (2002).
Zha, D. X., Eipper, A. & Reetz, M. T. Assembly of designed oligonucleotides as an efficient method for gene recombination: a new tool in directed evolution. Chembiochem 4, 34–39 (2003).
Crameri, A., Whitehorn, E. A., Tate, E. & Stemmer, W. P. C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14, 315–319 (1996).
Crameri, A., Raillard, S. A., Bermudez, E. & Stemmer, W. P. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 (1998).
Romanini, D. W., Peralta-Yahya, P., Mondol, V. & Cornish, V. W. A. Heritable recombination system for synthetic Darwinian evolution in yeast. ACS Synth. Biol. 1, 602–609 (2012).
Sieber, V., Martinez, C. A. & Arnold, F. H. Libraries of hybrid proteins from distantly related sequences. Nat. Biotechnol. 19, 456–460 (2001).
Ostermeier, M., Shim, J. H. & Benkovic, S. J. A combinatorial approach to hybrid enzymes independent of DNA homology. Nat. Biotechnol. 17, 1205–1209 (1999).
Bittker, J. A., Le, B. V., Liu, J. M. & Liu, D. R. Directed evolution of protein enzymes using nonhomologous random recombination. Proc. Natl Acad. Sci. USA 101, 7011–7016 (2004).
Voigt, C. A., Martinez, C., Wang, Z. G., Mayo, S. L. & Arnold, F. H. Protein building blocks preserved by recombination. Nat. Struct. Biol. 9, 553–558 (2002).
Hiraga, K. & Arnold, F. H. General method for sequence-independent site-directed chimeragenesis. J. Mol. Biol. 330, 287–296 (2003).
Kolkman, J. A. & Stemmer, W. P. C. Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 19, 423–428 (2001).
Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K. & Pease, L. R. Engineering hybrid genes without the use of restriction enzymes: gene-splicing by overlap extension. Gene 77, 61–68 (1989).
Gillam, E. M. J. Directed Evolution Library Creation (Springer, 2014). This book is an excellent resource for comparing and choosing between genetic diversification methods as well as for successfully executing library generation protocols.
You, L. & Arnold, F. H. Directed evolution of subtilisin E in Bacillus subtilis to enhance total activity in aqueous dimethylformamide. Protein Eng. 9, 77–83 (1996).
Heim, R., Prasher, D. C. & Tsien, R. Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl Acad. Sci. USA 91, 12501–12504 (1994).
Kralj, J. M., Douglass, A. D., Hochbaum, D. R., Maclaurin, D. & Cohen, A. E. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods 9, 90–130 (2012).
Cali, J. J. et al. Luminogenic cytochrome P450 assays. Expert Opin. Drug Metab. Toxicol. 2, 629–645 (2006).
Ostafe, R., Prodanovic, R., Lloyd Ung, W., Weitz, D. A. & Fischer, R. A high-throughput cellulase screening system based on droplet microfluidics. Biomicrofluidics 8, 041102 (2014).
Gupta, R. D. et al. Directed evolution of hydrolases for prevention of G-type nerve agent intoxication. Nat. Chem. Biol. 7, 120–125 (2011).
Giger, L. et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat. Chem. Biol. 9, 494–498 (2013).
Goddard, J. P. & Reymond, J. L. Enzyme assays for high-throughput screening. Curr. Opin. Biotechnol. 15, 314–322 (2004).
Fields, S. & Song, O. K. A novel genetic system to detect protein–protein interactions. Nature 340, 245–246 (1989).
Baker, K. et al. Chemical complementation: a reaction-independent genetic assay for enzyme catalysis. Proc. Natl Acad. Sci. USA 99, 16537–16542 (2002).
Lin, H. N., Tao, H. Y. & Cornish, V. W. Directed evolution of a glycosynthase via chemical complementation. J. Am. Chem. Soc. 126, 15051–15059 (2004).
Peralta-Yahya, P., Carter, B. T., Lin, H. N., Tao, H. Y. & Comish, V. W. High-throughput selection for cellulase catalysts using chemical complementation. J. Am. Chem. Soc. 130, 17446–17452 (2008).
Swe, P. M. et al. Targeted mutagenesis of the Vibrio fischeri flavin reductase FRase I to improve activation of the anticancer prodrug CB1954. Biochem. Pharmacol. 84, 775–783 (2012).
Sengupta, D., Lin, H. N., Goldberg, S. D., Mahal, J. J. & Cornish, V. W. Correlation between catalytic efficiency and the transcription read-out in chemical complementation: a general assay for enzyme catalysis. Biochemistry 43, 3570–3581 (2004).
Fulwyler, M. J. Electronic separation of biological cells by volume. Science 150, 910–911 (1965).
Shapiro, H. M. Practical Flow Cytometry (Wiley-Liss, 2003).
Boder, E. T. & Wittrup, K. D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997). This paper describes the invention of yeast display protein libraries for screening protein–protein interactions and serves as the foundation for many other cell surface display methods.
Santoro, S. W. & Schultz, P. G. Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA 99, 4185–4190 (2002).
Wang, J. D., Herman, C., Tipton, K. A., Gross, C. A. & Weissman, J. S. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell 111, 1027–1039 (2002).
Peck, S. H., Chen, I. & Liu, D. R. Directed evolution of a small-molecule-triggered intein with improved splicing properties in mammalian cells. Chem. Biol. 18, 619–630 (2011).
Rajpal, A. et al. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc. Natl Acad. Sci. USA 102, 8466–8471 (2005).
Wang, X. X., Cho, Y. K. & Shusta, E. V. Mining a yeast library for brain endothelial cell-binding antibodies. Nat. Methods 4, 143–145 (2007).
Chen, I., Dorr, B. M. & Liu, D. R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl Acad. Sci. USA 108, 11399–11404 (2011).
Qu, Z. et al. Immobilization of actively thromboresistant assemblies on sterile blood-contacting surfaces. Adv. Healthc. Mater. 3, 30–35 (2014).
Shi, J. H. et al. Engineered red blood cells as carriers for systemic delivery of a wide array of functional probes. Proc. Natl Acad. Sci. USA 111, 10131–10136 (2014).
Ling, J. J., Policarpo, R. L., Rabideau, A. E., Liao, X. & Pentelute, B. L. Protein thioester synthesis enabled by sortase. J. Am. Chem. Soc. 134, 10749–10752 (2012).
McCluskey, A. J. & Collier, R. J. Receptor-directed chimeric toxins created by sortase-mediated protein fusion. Mol. Cancer Ther. 12, 2273–2281 (2013).
Policarpo, R. L. et al. Flow-based enzymatic ligation by sortase A. Angew. Chem. Int. Ed Engl. 53, 9203–9208 (2014).
Swee, L. K., Lourido, S., Bell, G. W., Ingram, J. R. & Ploegh, H. L. One-step enzymatic modification of the cell surface redirects cellular cytotoxicity and parasite tropism. ACS Chem. Biol. (2014).
Dorr, B. M., Ham, H. O., An, C., Chaikof, E. L. & Liu, D. R. Reprogramming the specificity of sortase enzymes. Proc. Natl Acad. Sci. USA 111, 13343–13348 (2014).
Yi, L. et al. Engineering of TEV protease variants by yeast ER sequestration screening (YESS) of combinatorial libraries. Proc. Natl Acad. Sci. USA 110, 7229–7234 (2013).
Tawfik, D. S. & Griffiths, A. D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16, 652–656 (1998). The authors of this paper developed IVC as a platform for directed evolution. This study describes a selection for methyltransferases within water–oil emulsion droplets.
Bernath, K. et al. In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Anal. Biochem. 325, 151–157 (2004).
Agresti, J. J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl Acad. Sci. USA 107, 4004–4009 (2010).
Scott, D. J. & Plückthun, A. Direct molecular evolution of detergent-stable G protein-coupled receptors using polymer encapsulated cells. J. Mol. Biol. 425, 662–677 (2013).
Fischlechner, M. et al. Evolution of enzyme catalysts caged in biomimetic gel-shell beads. Nat. Chem. 6, 791–796 (2014). In this study, polyelectrolyte shells served as in vitro compartments for screening by flow cytometry.
Bessette, P. H., Rice, J. J. & Daugherty, P. S. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng. Des. Sel. 17, 731–739 (2004).
Mccafferty, J., Griffiths, A. D., Winter, G. & Chiswell, D. J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990). In this pioneering study, phage display is demonstrated as a powerful technique to select high-affinity antibody fragments. This paper also nicely illustrates the guiding principles of related binding enrichments.
Clackson, T., Hoogenboom, H. R., Griffiths, A. D. & Winter, G. Making antibody fragments using phage display libraries. Nature 352, 624–628 (1991).
Scott, J. K. & Smith, G. P. Searching for peptide ligands with an epitope library. Science 249, 386–390 (1990).
Becker, D. M. & Guarente, L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 194, 182–187 (1991).
Dower, W. J., Miller, J. F. & Ragsdale, C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145 (1988).
Hanes, J. & Pluckthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl Acad. Sci. USA 94, 4937–4942 (1997).
Wilson, D. S., Keefe, A. D. & Szostak, J. W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl Acad. Sci. USA 98, 3750–3755 (2001).
Amstutz, P. et al. In vitro selection for catalytic activity with ribosome display. J. Am. Chem. Soc. 124, 9396–9403 (2002).
Seelig, B. & Szostak, J. W. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature 448, 828–831 (2007).
Orencia, M. C., Yoon, J. S., Ness, J. E., Stemmer, W. P. & Stevens, R. C. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat. Struct. Biol. 8, 238–242 (2001).
Palmer, A. C. & Kishony, R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet. 14, 243–248 (2013).
Liu, D. R., Magliery, T. J., Pasternak, M. & Schultz, P. G. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc. Natl Acad. Sci. USA 94, 10092–10097 (1997). This groundbreaking study on genetic code expansion exemplifies how selectable antibiotic resistance markers can form the basis for a range of in vivo selections.
Santoro, S. W., Wang, L., Herberich, B., King, D. S. & Schultz, P. G. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 20, 1044–1048 (2002).
Gaj, T., Mercer, A. C., Gersbach, C. A., Gordley, R. M. & Barbas, C. F. 3rd Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl Acad. Sci. USA 108, 498–503 (2011).
Young, E. M., Tong, A., Bui, H., Spofford, C. & Alper, H. S. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc. Natl Acad. Sci. USA 111, 131–136 (2014). This study uses an auxotroph complementation strategy to select for sugar transporters that selectively uptake xylose from culture media.
Lee, S. M., Jellison, T. & Alper, H. S. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 78, 5708–5716 (2012).
Worsdorfer, B., Woycechowsky, K. J. & Hilvert, D. Directed evolution of a protein container. Science 331, 589–592 (2011).
Takeuchi, R., Choi, M. & Stoddard, B. L. Redesign of extensive protein–DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc. Natl Acad. Sci. USA 111, 4061–4066 (2014).
Ghadessy, F. J., Ong, J. L. & Holliger, P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl Acad. Sci. USA 98, 4552–4557 (2001).
Ramsay, N. et al. CyDNA: synthesis and replication of highly Cy-dye substituted DNA by an evolved polymerase. J. Am. Chem. Soc. 132, 5096–5104 (2010).
d'Abbadie, M. et al. Molecular breeding of polymerases for amplification of ancient DNA. Nat. Biotechnol. 25, 939–943 (2007).
Ellefson, J. W. et al. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat. Biotechnol. 32, 97–101 (2014). The authors of this paper evolved enzymes within IVCs by linking the desired phenotype to the expression of Taq polymerase within E. coli . Taq can then be used in PCR to amplify the DNA encoding active library members within the emulsion droplet.
Meyer, A. J., Ellefson, J. W. & Ellington, A. D. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry. ACS Synth. Biol. http:///dx.doi.org/10.1021/sb500299c (2014).
Badran, A. H. & Liu, D. R. In vivo continuous directed evolution. Curr. Opin. Chem. Biol. 24, 1–10 (2015).
Lenski, R. E. & Travisano, M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl Acad. Sci. USA 91, 6808–6814 (1994).
Toprak, E. et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 44, 101–105 (2012).
Muller, M. M. et al. Directed evolution of a model primordial enzyme provides insights into the development of the genetic code. PLoS Genet. 9, e1003187 (2013).
Camps, M., Naukkarinen, J., Johnson, B. P. & Loeb, L. A. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc. Natl Acad. Sci. USA 100, 9727–9732 (2003).
Bull, J. J. et al. Exceptional convergent evolution in a virus. Genetics 147, 1497–1507 (1997).
Wichman, H. A., Wichman, J. & Bull, J. J. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics 170, 19–31 (2005).
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011). This study establishes a technological platform for the continuous evolution of biomolecules by linking the phage life cycle to the desired enzymatic activity.
Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A. & Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 10, 216–222 (2014).
Dickinson, B. C., Packer, M. S., Badran, A. H. & Liu, D. R. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nat. Commun. 5, 5352 (2014).
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008).
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science 329, 309–313 (2010).
Joh, N. H. et al. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524 (2014).
King, N. P. et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108 (2014).
Song, W. J. & Tezcan, F. A. A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346, 1525–1528 (2014).
Karanicolas, J. et al. A de novo protein binding pair by computational design and directed evolution. Mol. Cell 42, 250–260 (2011).
Patel, S. C. & Hecht, M. H. Directed evolution of the peroxidase activity of a de novo-designed protein. Protein Eng. Des. Sel. 25, 445–452 (2012).
Khersonsky, O. et al. Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution. J. Mol. Biol. 407, 391–412 (2011).
Rothlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008). This paper describes the computational design of a Kemp elimination catalyst. Subsequent screening yielded improved catalysts for a reaction that is not known to be performed by natural enzymes.
Savile, C. K. et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309 (2010).
Lutz, S. & Patrick, W. M. Novel methods for directed evolution of enzymes: quality, not quantity. Curr. Opin. Biotechnol. 15, 291–297 (2004).
Becker, S. et al. Single-cell high-throughput screening to identify enantioselective hydrolytic enzymes. Angew. Chem. Int. Ed Engl. 47, 5085–5088 (2008).
Lipovsek, D. et al. Selection of horseradish peroxidase variants with enhanced enantioselectivity by yeast surface display. Chem. Biol. 14, 1176–1185 (2007).
Piotukh, K. et al. Directed evolution of sortase A mutants with altered substrate selectivity profiles. J. Am. Chem. Soc. 133, 17536–17539 (2011).
Acknowledgements
This work was supported by the US Defense Advanced Research Projects Agency grants DARPA HR0011-11-2-0003 and DARPA N66001-12-C-4207, the US National Institutes of Health (NIH)/National Institute of General Medical Sciences (NIGMS) (grant R01 GM095501) and the Howard Hughes Medical Institute (HHMI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Natural selection
-
A process by which individuals with the highest reproductive fitness pass on their genetic material to their offspring, thus maintaining and enriching heritable traits that are adaptive to the natural environment.
- Artificial selection
-
(Also known as selective breeding). A process by which human intervention in the reproductive cycle imposes a selection pressure for phenotypic traits desired by the breeder.
- Libraries
-
Diverse populations of DNA fragments that are subject to downstream screening and selection.
- Library size
-
The number variants that are subjected to screening and selection. Library sizes are limited by molecular cloning protocols and/or by host transformation efficiency.
- Focused mutagenesis
-
A strategy of diversification that introduces mutations at DNA regions expected to influence protein activity.
- Random mutagenesis
-
A strategy of diversification that introduces mutations in an unbiased manner throughout the entire gene.
- Mutational spectrum
-
The frequency of each specific type of transition and transversion. The evenness of this spectrum allows more thorough sampling of sequence space.
- Transformation
-
The process by which a cell directly acquires a foreign DNA molecule. A number of protocols allow high-efficiency transformation of microorganisms through treatments with ionic buffers, heat shock or electroporation.
- Neutral drift
-
A process that occurs in the presence of a purifying selection pressure to eliminate deleterious mutations. This is in contrast to genetic drift, a process by which mutations fluctuate in frequency in the absence of selection pressure.
- Degenerate codons
-
Codons constructed with a mixed population of nucleotides at a given position, thus sampling all possible amino acids within the constructed libraries. The most popular examples are NNK and NNS (where N can be any of the four nucleotides, K can be G or T, and S can be G or C).
- Epistatic interactions
-
Non-additive effects between mutations (for example, mutational synergy or synthetic lethality). As a result, the sequential acquisition of mutations is not always equivalent to mutational co-occurrence.
- Homologous recombination
-
A process by which separate pieces of DNA swap genetic material, guided by the annealing of complementary DNA fragments.
- Passenger mutations
-
(Also known as hitchhiker mutations). Unnecessary mutations that are enriched in a population owing to co-occurrence with a highly beneficial linked mutation.
- Transduction
-
The process by which a viral vector delivers a foreign DNA molecule to a cellular host.
- Evolutionary potential
-
The capacity of a protein to take on new functions through evolution. High thermostability allows for necessary but destabilizing mutations, and functional diversity of homologues is a demonstration of previous evolution in nature.
- Surrogate substrates
-
Substrate analogues that are permissive of enzymatic conversion but that, upon catalysis, exhibit chemical rearrangements that lead to altered optical properties, including visible colour, relief of fluorophore quenching, shifted fluorophore excitation or emission, and downstream chemiluminescence.
- Fluorescence-activated cell sorting
-
(FACS). A flow cytometry method in which an aqueous suspension of cells or cell-like compartments is measured for fluorescence (often at multiple wavelengths) one cell at a time and subsequently separated based on a fluorescence threshold.
- Negative screen
-
A screening method that involves depletion of an undesired phenotype.
- Positive screening
-
Enrichment for a desired activity such as improved kinetics, tolerance to unnatural conditions and acceptance of new substrates.
- Transformation bottleneck
-
The efficiency at which DNA library members are transferred into the host organism, thus restricting the number of variants that can be assessed by in vivo selection and screening.
- Auxotroph complementation
-
The ability of functional library members to resolve a metabolic defect in the host, leading to replication of DNA that encodes active library members.
Rights and permissions
About this article
Cite this article
Packer, M., Liu, D. Methods for the directed evolution of proteins. Nat Rev Genet 16, 379–394 (2015). https://doi.org/10.1038/nrg3927
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3927
This article is cited by
-
A novel computationally engineered collagenase reduces the force required for tooth extraction in an ex-situ porcine jaw model
Journal of Biological Engineering (2023)
-
A biocatalytic platform for asymmetric alkylation of α-keto acids by mining and engineering of methyltransferases
Nature Communications (2023)
-
ACIDES: on-line monitoring of forward genetic screens for protein engineering
Nature Communications (2023)
-
Engineered bacterial orthogonal DNA replication system for continuous evolution
Nature Chemical Biology (2023)
-
Olfactory marker protein contains a leucine-rich domain in the Ω-loop important for nuclear export
Molecular Brain (2022)